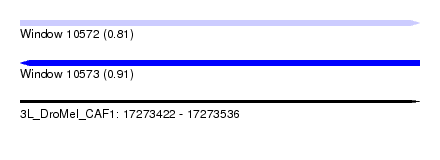
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,273,422 – 17,273,536 |
| Length | 114 |
| Max. P | 0.905191 |

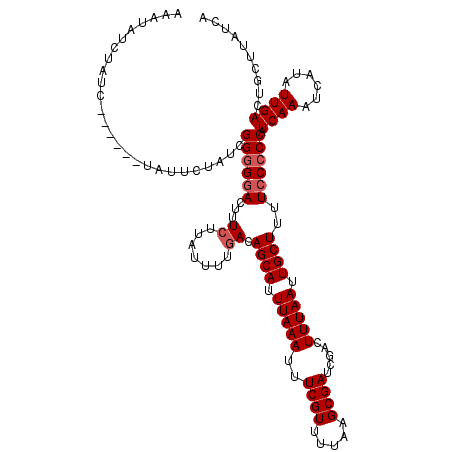
| Location | 17,273,422 – 17,273,536 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -18.91 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17273422 114 + 23771897 UGAUAAGCAGUCAAUAUGAUUUGAUGGGGGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAUAAAGUUCCCCGAUAUAAUA------GAUACAUAUUU .(((.....)))(((((((((((((.((((((.((((.(((((.(((...........)))..))))).))))...............)))))).))....))------))).)))))). ( -20.89) >DroSec_CAF1 156923 114 + 1 UGAUAAGCAGUCAAUAUGAUUUGAUGGGGGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAGAAAGUCCCCCGAUAGAAUA------GAUAGAUAUUU .........(((.((...((((..(((((((..((((.(((((.(((...........)))..))))).)))).((.......))...)))))))...)))).------.)).))).... ( -21.40) >DroSim_CAF1 151992 114 + 1 UGAUAAGCAGUCAAUAUGAUUUGAUGGGGGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAGAAAGUCCCCCGAUAGAAUA------GAUAGAUAUUU .........(((.((...((((..(((((((..((((.(((((.(((...........)))..))))).)))).((.......))...)))))))...)))).------.)).))).... ( -21.40) >DroEre_CAF1 159789 119 + 1 UGAUAAGCAGUCAAUAUGAUUUGAUGG-GGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAGAAAGUCCCCCCACAGAAUAGAUAUAGAUAGAUAUUU .........(((.((((.((((..(((-((...((((.(((((.(((...........)))..))))).)))).((.......))......)))))..))))..)))).)))........ ( -20.00) >DroYak_CAF1 164680 114 + 1 UGAUAAGCAGUCAAUAUGAUUUGAUGGGGGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAGAAAGUCCCCCGAUAGAAUA------CAUAGAUAUUU ((((.....)))).((((((((..(((((((..((((.(((((.(((...........)))..))))).)))).((.......))...)))))))...)))).------))))....... ( -23.40) >consensus UGAUAAGCAGUCAAUAUGAUUUGAUGGGGGAAAAGCAAUUAAAGUCGAUCGCUUAAAACGAAAUUUAAAUGCUGUCAAAAUAAGAAAGUCCCCCGAUAGAAUA______GAUAGAUAUUU .........(((((......)))))((((((..((((.(((((.(((...........)))..))))).))))...............)))))).......................... (-18.91 = -18.95 + 0.04)

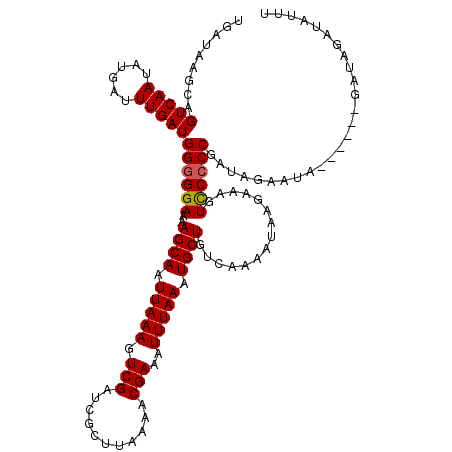
| Location | 17,273,422 – 17,273,536 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -20.08 |
| Consensus MFE | -16.70 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17273422 114 - 23771897 AAAUAUGUAUC------UAUUAUAUCGGGGAACUUUAUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCCCCCAUCAAAUCAUAUUGACUGCUUAUCA ......(((..------.........((((((..(((......))).((((.(((((..((((.....)))).....))))).)))).))))))..((((......)))).)))...... ( -17.20) >DroSec_CAF1 156923 114 - 1 AAAUAUCUAUC------UAUUCUAUCGGGGGACUUUCUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCCCCCAUCAAAUCAUAUUGACUGCUUAUCA ...........------.........((((((...((.......)).((((.(((((..((((.....)))).....))))).))))..)))))).((((......)))).......... ( -20.70) >DroSim_CAF1 151992 114 - 1 AAAUAUCUAUC------UAUUCUAUCGGGGGACUUUCUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCCCCCAUCAAAUCAUAUUGACUGCUUAUCA ...........------.........((((((...((.......)).((((.(((((..((((.....)))).....))))).))))..)))))).((((......)))).......... ( -20.70) >DroEre_CAF1 159789 119 - 1 AAAUAUCUAUCUAUAUCUAUUCUGUGGGGGGACUUUCUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCC-CCAUCAAAUCAUAUUGACUGCUUAUCA .........((.((((..(((..((((((..(...((.......)).((((.(((((..((((.....)))).....))))).)))))..))-))))..))).)))).)).......... ( -20.80) >DroYak_CAF1 164680 114 - 1 AAAUAUCUAUG------UAUUCUAUCGGGGGACUUUCUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCCCCCAUCAAAUCAUAUUGACUGCUUAUCA ..........(------((.......((((((...((.......)).((((.(((((..((((.....)))).....))))).))))..)))))).((((......)))).)))...... ( -21.00) >consensus AAAUAUCUAUC______UAUUCUAUCGGGGGACUUUCUUAUUUUGACAGCAUUUAAAUUUCGUUUUAAGCGAUCGACUUUAAUUGCUUUUCCCCCAUCAAAUCAUAUUGACUGCUUAUCA ..........................((((((...((.......)).((((.(((((..((((.....)))).....))))).))))..)))))).((((......)))).......... (-16.70 = -17.30 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:40 2006