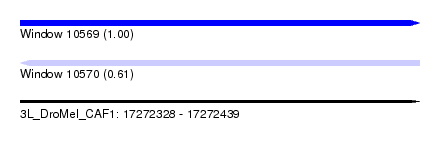
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,272,328 – 17,272,439 |
| Length | 111 |
| Max. P | 0.998913 |

| Location | 17,272,328 – 17,272,439 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -36.94 |
| Consensus MFE | -20.62 |
| Energy contribution | -21.56 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17272328 111 + 23771897 GGACGUGAGGAAGGGCAGGGAAAGGA---------CCUGAGACCUGGGCAAAAAGCAAGCGAGGGGUGAAUAGCUUUAAAGCUGAUGCAUAAAAUAUUGCGUAUACGCCCCAUUAUGCCA ...(....)..(((.((((.......---------))))...))).((((....(....)..((((((..(((((....)))))(((((........)))))...))))))....)))). ( -37.00) >DroSec_CAF1 155844 111 + 1 AGACGUGAGGAGGGGCAGGGGAAGGA---------CCUGAGACCUGGGCAAAUAGCAAGCGAGGGGUGAAUAGCUUUAAAGCUGAUGCAUAAAAUAUUGCGUAUACGCCCCAUUAUGCCA ...(....).(((..((((.......---------))))...))).((((....(....)..((((((..(((((....)))))(((((........)))))...))))))....)))). ( -37.50) >DroSim_CAF1 150881 111 + 1 GGACGUGAGGAGGAGCAGGGGAAGGA---------CCUGAGACCUGGGCAAAUAGCAAGCGAGGGGUGAAUAGCUUUAAAGCUGAUGCAUAAAAUAUUGCGUAUACGCCCCAUUAUGCCA ...(....).(((..((((.......---------))))...))).((((....(....)..((((((..(((((....)))))(((((........)))))...))))))....)))). ( -36.40) >DroEre_CAF1 158684 85 + 1 GGAUGUGAGAAGGGGACGG-----------------------------------GCAGCGGCAGGGGCAGGAGCUUUAAAGCUGAUGCAUAAUAUAUUGCGUAUACGCCCCAUUAUGCCC ...........(....)((-----------------------------------(((......(((((...((((....)))).(((((........)))))....)))))....))))) ( -29.00) >DroYak_CAF1 163499 114 + 1 GGACGUGAAUGGGAGAAGGA--GGCUCCUCCUCCUCCUG----CUGGGUAAAAAGCAUGCGAGGGGUGAAUAGCUUUAAAGCUGAUGCAUAAUAUAUUGCGUAUACGCCCCAUUAUGCCC ((.(((((..(((((.((((--(....))))).)))))(----((........)))......((((((..(((((....)))))(((((........)))))...)))))).))))))). ( -44.80) >consensus GGACGUGAGGAGGGGCAGGG_AAGGA_________CCUGAGACCUGGGCAAAAAGCAAGCGAGGGGUGAAUAGCUUUAAAGCUGAUGCAUAAAAUAUUGCGUAUACGCCCCAUUAUGCCA ((.(((((...........................((........))...............((((((..(((((....)))))(((((........)))))...)))))).))))))). (-20.62 = -21.56 + 0.94)

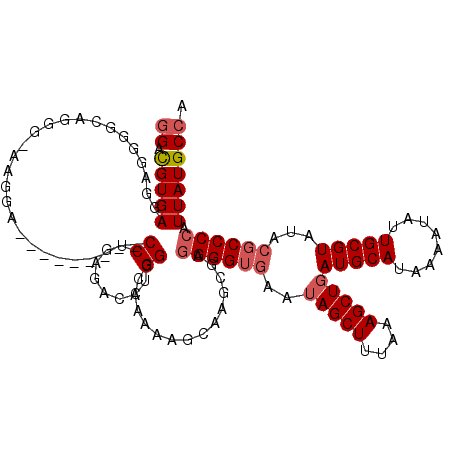

| Location | 17,272,328 – 17,272,439 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.58 |
| Mean single sequence MFE | -26.89 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17272328 111 - 23771897 UGGCAUAAUGGGGCGUAUACGCAAUAUUUUAUGCAUCAGCUUUAAAGCUAUUCACCCCUCGCUUGCUUUUUGCCCAGGUCUCAGG---------UCCUUUCCCUGCCCUUCCUCACGUCC .((((....((((.(.....(((........)))...((((....))))...).)))).....))))........(((...((((---------.......)))).....)))....... ( -21.90) >DroSec_CAF1 155844 111 - 1 UGGCAUAAUGGGGCGUAUACGCAAUAUUUUAUGCAUCAGCUUUAAAGCUAUUCACCCCUCGCUUGCUAUUUGCCCAGGUCUCAGG---------UCCUUCCCCUGCCCCUCCUCACGUCU .(((......(((((.(((.((((.............((((....)))).............))))))).)))))(((...((((---------.......))))..)))......))). ( -24.07) >DroSim_CAF1 150881 111 - 1 UGGCAUAAUGGGGCGUAUACGCAAUAUUUUAUGCAUCAGCUUUAAAGCUAUUCACCCCUCGCUUGCUAUUUGCCCAGGUCUCAGG---------UCCUUCCCCUGCUCCUCCUCACGUCC .(((......(((((.(((.((((.............((((....)))).............))))))).)))))(((...((((---------.......))))..)))......))). ( -24.37) >DroEre_CAF1 158684 85 - 1 GGGCAUAAUGGGGCGUAUACGCAAUAUAUUAUGCAUCAGCUUUAAAGCUCCUGCCCCUGCCGCUGC-----------------------------------CCGUCCCCUUCUCACAUCC (((((....(((((......(((........)))...((((....))))...)))))......)))-----------------------------------))................. ( -25.10) >DroYak_CAF1 163499 114 - 1 GGGCAUAAUGGGGCGUAUACGCAAUAUAUUAUGCAUCAGCUUUAAAGCUAUUCACCCCUCGCAUGCUUUUUACCCAG----CAGGAGGAGGAGGAGCC--UCCUUCUCCCAUUCACGUCC ((((((((((..(((....)))....))))))))...((((....)))).......)).....((((........))----))((((((((((....)--)))))))))........... ( -39.00) >consensus UGGCAUAAUGGGGCGUAUACGCAAUAUUUUAUGCAUCAGCUUUAAAGCUAUUCACCCCUCGCUUGCUAUUUGCCCAGGUCUCAGG_________UCCUU_CCCUGCCCCUCCUCACGUCC .((((....((((.......(((........)))...((((....)))).....)))).....))))..................................................... (-15.20 = -15.24 + 0.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:37 2006