
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,271,672 – 17,271,869 |
| Length | 197 |
| Max. P | 0.999757 |

| Location | 17,271,672 – 17,271,792 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921698 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271672 120 + 23771897 UUUCGGAGUUGCCGGGAAAAACUCUGUGAAAAAUAGAGCGCCGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU (..(((.....)))..)....((((((.....))))))((((((((.((((((....)))))).....)))))).))...(((.........)))...(((((........))))).... ( -31.10) >DroSec_CAF1 155188 120 + 1 UUUCGGAGUUGCCGGGAAAAACUCUACGAAAAAUAGAGCGCAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU .....(((((((((..(.....((((.(((....(((((.(((....))).)))))..)))...)))))..)))).....(((.........)))..))))).................. ( -26.40) >DroSim_CAF1 150221 120 + 1 UUUCGGAGUUGCCGGGAAAAACUCUGCGAAAAAUAGAGCGCAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU (..(((.....)))..).......((((((....(((((.(((....))).)))))..)))....((.(((...(((....)))...))).)))))..(((((........))))).... ( -29.10) >DroEre_CAF1 158032 120 + 1 UUUCGGAGUUGCCGGGGAAAACUCUGGGAAAAAUAGAGCGCAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU ..((((((((.((((((....))))))(((....(((((.(((....))).)))))..))).....))))))))......(((.........)))...(((((........))))).... ( -32.00) >DroYak_CAF1 162840 120 + 1 UUUCGGAGUUGCCGGCGAAAACUCGGUGAAAAAUAGAGCGCAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU .....(((((..((((((((.((...((((....(((((.(((....))).)))))..))))...)).))))).(((....))).......)))...))))).................. ( -30.00) >consensus UUUCGGAGUUGCCGGGAAAAACUCUGCGAAAAAUAGAGCGCAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAU .....(((((..(((............(((....(((((.(((....))).)))))..)))......((((((........))))))....)))...))))).................. (-25.10 = -25.30 + 0.20)

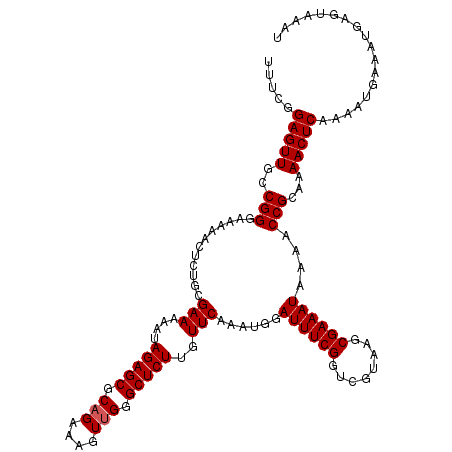

| Location | 17,271,712 – 17,271,832 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.36 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271712 120 + 23771897 CCGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU ((((((.((((((....)))))).....))))))(((....))).....((((((((.(((((........)))))...))))))))((((((......)))))).(((....))).... ( -35.50) >DroSec_CAF1 155228 120 + 1 CAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU ..((((.((((((....)))))).....))))(((.(...((.......((((((((.(((((........)))))...))))))))((((((......)))))).))....).)))... ( -32.30) >DroSim_CAF1 150261 120 + 1 CAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU ..((((.((((((....)))))).....))))(((.(...((.......((((((((.(((((........)))))...))))))))((((((......)))))).))....).)))... ( -32.30) >DroEre_CAF1 158072 120 + 1 CAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUUAGCACUCCUGCCUUU ...((((((.(((.(((..........((((((........))))))..((((((((.(((((........)))))...)))))))).)))..)))...)))))).(((....))).... ( -31.20) >DroYak_CAF1 162880 120 + 1 CAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUUAGCACUCCUGCCUUU ...((((((.(((.(((..........((((((........))))))..((((((((.(((((........)))))...)))))))).)))..)))...)))))).(((....))).... ( -31.20) >consensus CAGAAAGUUGGGCUCUUGUUCAAAUGGAUUUCGGUCGUAAGCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU ..((((.((((((....)))))).....))))(((.(...((.......((((((((.(((((........)))))...))))))))((((((......)))))).))....).)))... (-31.60 = -31.36 + -0.24)

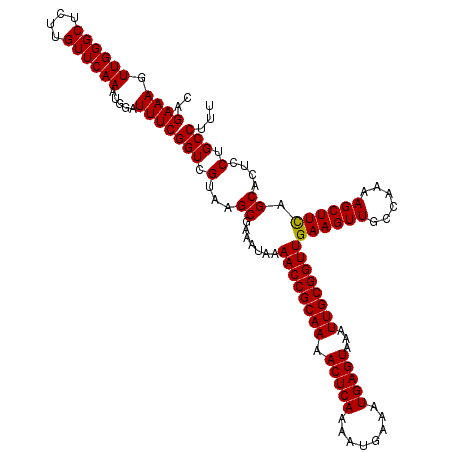
| Location | 17,271,712 – 17,271,832 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -33.00 |
| Energy contribution | -32.76 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271712 120 - 23771897 AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCGG ..((((.(((((((..(.....)..)).)))))((((((((...(((((........))))).)))))))).......))))....((((((....(((.....))).......)))))) ( -33.30) >DroSec_CAF1 155228 120 - 1 AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCUG ...(((.(((((((..(.....)..)).)))))((((((((...(((((........))))).))))))))..((((((........))))))..............))).......... ( -32.80) >DroSim_CAF1 150261 120 - 1 AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCUG ...(((.(((((((..(.....)..)).)))))((((((((...(((((........))))).))))))))..((((((........))))))..............))).......... ( -32.80) >DroEre_CAF1 158072 120 - 1 AAAGGCAGGAGUGCUAAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCUG ...(((.((((((((((.....))))).)))))((((((((...(((((........))))).))))))))..((((((........))))))..............))).......... ( -33.30) >DroYak_CAF1 162880 120 - 1 AAAGGCAGGAGUGCUAAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCUG ...(((.((((((((((.....))))).)))))((((((((...(((((........))))).))))))))..((((((........))))))..............))).......... ( -33.30) >consensus AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGCUUACGACCGAAAUCCAUUUGAACAAGAGCCCAACUUUCUG ...(((.((((((((((.....))))).)))))((((((((...(((((........))))).))))))))..((((((........))))))..............))).......... (-33.00 = -32.76 + -0.24)

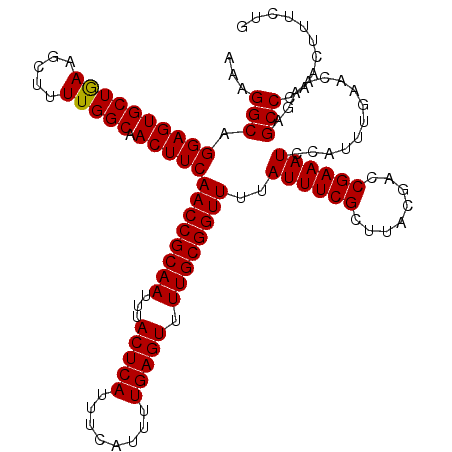
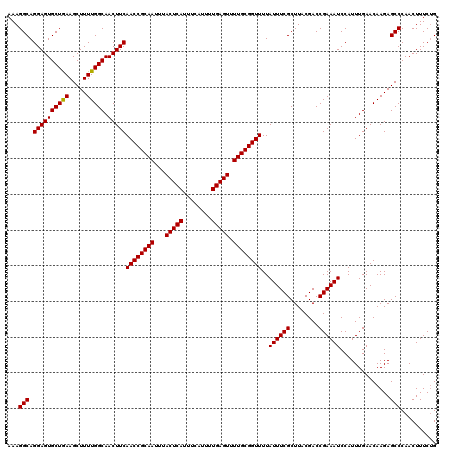
| Location | 17,271,752 – 17,271,869 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.08 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271752 117 + 23771897 GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU-UUUGGAAUGUGUGGUUUUC--GUUUUGCUUGGCCACAUC ((((((((((((((((..(((((........)))))........(((((((((......)))))))))..(((.......-...)))...))))))))).--)))))))........... ( -34.40) >DroSec_CAF1 155268 117 + 1 GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU-UUUGGAAUGUGUGGUUUUC--GUUUUGCUUGGCCACAUC ((((((((((((((((..(((((........)))))........(((((((((......)))))))))..(((.......-...)))...))))))))).--)))))))........... ( -34.40) >DroSim_CAF1 150301 117 + 1 GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU-UUUGGAAUGUGUGGUUUUC--GUUUUGCUUGGCCACAUC ((((((((((((((((..(((((........)))))........(((((((((......)))))))))..(((.......-...)))...))))))))).--)))))))........... ( -34.40) >DroEre_CAF1 158112 117 + 1 GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUUAGCACUCCUGCCUUU-UUUGGAAUGUGUGGUUUUU--GUUUUGCUUGGCCACAUC (((((((((((((((((.(((((........)))))...)))))))(((((((......)))))))((((.....((...-...))...))))....)))--)))))))........... ( -33.00) >DroYak_CAF1 162920 120 + 1 GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUUAGCACUCCUGCCUUUUUUUGGAAUGUGUGGUUUUUGUGUUUUGCUUGGCCACAUC ((((((((.((((((((.(((((........)))))...))))))))........((((((((...(((.(((...........))).)))..))))))))))))))))........... ( -35.50) >consensus GCGAAAUAAAACCGCAAAACUCAAAAUGAAAUGAGUAAAUUGCGGUUGAAGUUGCCAAAAGCUUCAGCACUCCUGCCUUU_UUUGGAAUGUGUGGUUUUC__GUUUUGCUUGGCCACAUC ((.......((((((((.(((((........)))))...))))))))((((((......)))))).))..(((...........)))..((((((((..............)))))))). (-32.32 = -32.08 + -0.24)

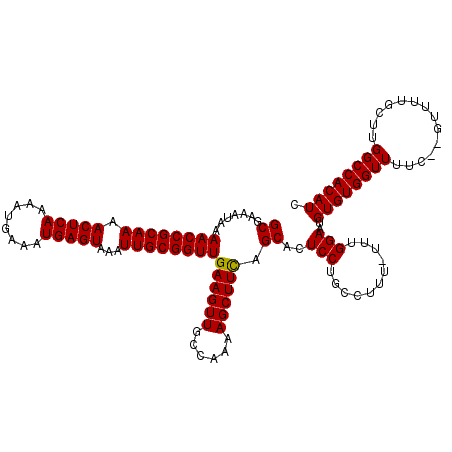
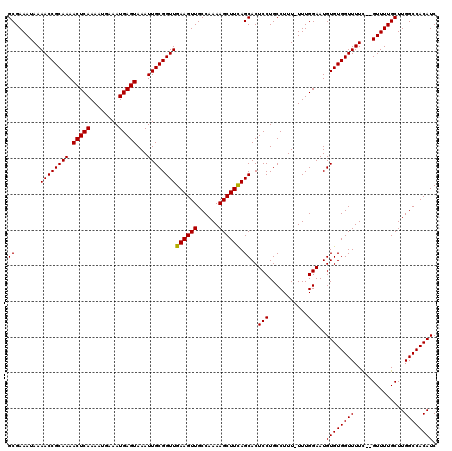
| Location | 17,271,752 – 17,271,869 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -32.41 |
| Consensus MFE | -30.57 |
| Energy contribution | -30.33 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.01 |
| SVM RNA-class probability | 0.999757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271752 117 - 23771897 GAUGUGGCCAAGCAAAAC--GAAAACCACACAUUCCAAA-AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ((.((.(((((((.....--(.....).((((((((...-.......)))))).))..)))..)))).)).))((((((((...(((((........))))).))))))))......... ( -33.40) >DroSec_CAF1 155268 117 - 1 GAUGUGGCCAAGCAAAAC--GAAAACCACACAUUCCAAA-AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ((.((.(((((((.....--(.....).((((((((...-.......)))))).))..)))..)))).)).))((((((((...(((((........))))).))))))))......... ( -33.40) >DroSim_CAF1 150301 117 - 1 GAUGUGGCCAAGCAAAAC--GAAAACCACACAUUCCAAA-AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ((.((.(((((((.....--(.....).((((((((...-.......)))))).))..)))..)))).)).))((((((((...(((((........))))).))))))))......... ( -33.40) >DroEre_CAF1 158112 117 - 1 GAUGUGGCCAAGCAAAAC--AAAAACCACACAUUCCAAA-AAAGGCAGGAGUGCUAAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ..(((((...........--.....))))).........-....((.((((((((((.....))))).)))))((((((((...(((((........))))).)))))))).......)) ( -30.99) >DroYak_CAF1 162920 120 - 1 GAUGUGGCCAAGCAAAACACAAAAACCACACAUUCCAAAAAAAGGCAGGAGUGCUAAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ..(((((..................)))))..............((.((((((((((.....))))).)))))((((((((...(((((........))))).)))))))).......)) ( -30.87) >consensus GAUGUGGCCAAGCAAAAC__GAAAACCACACAUUCCAAA_AAAGGCAGGAGUGCUGAAGCUUUUGGCAACUUCAACCGCAAUUUACUCAUUUCAUUUUGAGUUUUGCGGUUUUAUUUCGC ..(((((..................)))))..............((.((((((((((.....))))).)))))((((((((...(((((........))))).)))))))).......)) (-30.57 = -30.33 + -0.24)


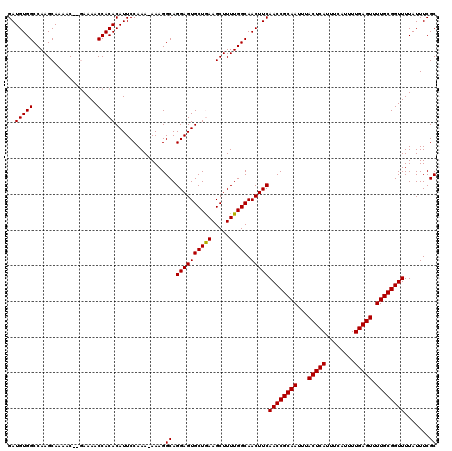
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:35 2006