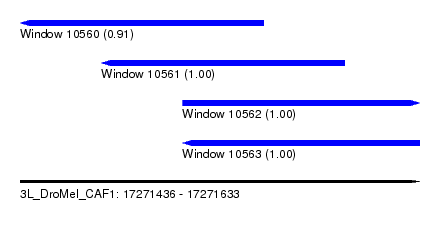
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,271,436 – 17,271,633 |
| Length | 197 |
| Max. P | 0.999949 |

| Location | 17,271,436 – 17,271,556 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.15 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271436 120 - 23771897 GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACGAAAACGAGUGAACGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.(((.(((....(((....(((..........))).)))....((((....((((((.....))))))((....)).....)))).....))).)))))))............... ( -27.40) >DroSec_CAF1 154952 120 - 1 GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACGAAAACGAGUGAACGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.(((.(((....(((....(((..........))).)))....((((....((((((.....))))))((....)).....)))).....))).)))))))............... ( -27.40) >DroSim_CAF1 149985 120 - 1 GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACGAAAACGAGUGAACGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.(((.(((....(((....(((..........))).)))....((((....((((((.....))))))((....)).....)))).....))).)))))))............... ( -27.40) >DroEre_CAF1 157801 114 - 1 GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAA------ACGAGUGAGCGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.(((((((.....(((.(.(((..........))).)..............((((((.....)))))))))..------...)))))))(......)))))............... ( -29.90) >DroYak_CAF1 162606 114 - 1 GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCCAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAA------ACGAGUGAGCGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.(((((((.....(((...(((...............)))...........((((((.....)))))))))..------...)))))))(......)))))............... ( -29.26) >consensus GCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACGAAAACGAGUGAACGAAUCAGCGGGCCAACUCAAAUGAAAC ((((.((((.....((((...(.(((..........))).)......))))....((((((.....)))))).............))))((.....))...))))............... (-24.90 = -25.10 + 0.20)



| Location | 17,271,476 – 17,271,596 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -36.16 |
| Consensus MFE | -34.72 |
| Energy contribution | -34.76 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271476 120 - 23771897 CUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACG .((((((((..(((((..((....))...(((((((((.((....((((.....)))))).))))))))).....))))))))............((((((.....)))))).))))).. ( -36.50) >DroSec_CAF1 154992 120 - 1 CUUUUUGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACG .((((((((..(((((..((....))...(((((((((.((....((((.....)))))).))))))))).....))))))))))))).......((((((.....))))))........ ( -37.70) >DroSim_CAF1 150025 120 - 1 CUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACG .((((((((..(((((..((....))...(((((((((.((....((((.....)))))).))))))))).....))))))))............((((((.....)))))).))))).. ( -36.50) >DroEre_CAF1 157838 117 - 1 CUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAA--- ..(((((((..(((((..((....))...(((((((((.((....((((.....)))))).))))))))).....))))))))............((((((.....)))))).))))--- ( -35.20) >DroYak_CAF1 162643 117 - 1 CUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCCAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAA--- .(((((((((......)))))))))....(((((((((.((....((((.....)))))).))))))))).........................((((((.....)))))).....--- ( -34.90) >consensus CUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAAGGCAAAAACUCAAACGGCGUUGGCAUGACGCCCGAAAACG ..(((((((...((((..((....))...(((((((((.((....((((.....)))))).))))))))).....)))).)))............((((((.....)))))).))))... (-34.72 = -34.76 + 0.04)


| Location | 17,271,516 – 17,271,633 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -32.34 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271516 117 + 23771897 UUGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCGAAAAGCGAACAGG---AGGCUAAAAGCAAAGGAUUUCCCAUUUUC (((((((((...(((((((.((.(((..........))))).)))))))(((((..((((((((......)))))))).)))))....---....))))))))).((.....))...... ( -34.90) >DroSec_CAF1 155032 117 + 1 UUGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCAAAAAGCGAACAGG---AGGCUAAAAGCAAAGGAUUUCCCAUUUUC (((((((((.(((((((((.((.(((..........))))).))))))))).((.((((((((..((.....))....)))))).)).---..))))))))))).((.....))...... ( -33.30) >DroSim_CAF1 150065 117 + 1 UUGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCGAAAAGCGAACAGG---AGGCUAAAAGCAAAGGAUUUCCCAUUUUC (((((((((...(((((((.((.(((..........))))).)))))))(((((..((((((((......)))))))).)))))....---....))))))))).((.....))...... ( -34.90) >DroEre_CAF1 157875 117 + 1 UUGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCGAAAAGCGAACAGG---AGGCUAAAAGCAAAGGAUUUCCCAUUUUC (((((((((...(((((((.((.(((..........))))).)))))))(((((..((((((((......)))))))).)))))....---....))))))))).((.....))...... ( -34.90) >DroYak_CAF1 162680 120 + 1 UGGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCGAAAAGCGAACAGGAGGAGGCCAAAAGCAAAGGAUUUCCCAUUUUC ((((((((....(((((((.((.(((..........))))).)))))))(((((..((((((((......)))))))).))))).....))))))))........((.....))...... ( -36.00) >consensus UUGCUUUUAUAAUUGCAUGUCGACUCUUAAUGAGCUGGGCGCCAUGCAAUUUGUACUUUCGCUGAGCAAACAGCGAAAAGCGAACAGG___AGGCUAAAAGCAAAGGAUUUCCCAUUUUC (((((((((...(((((((.((.(((..........))))).)))))))(((((..((((((((......)))))))).)))))...........))))))))).((.....))...... (-32.34 = -32.94 + 0.60)



| Location | 17,271,516 – 17,271,633 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.97 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -35.94 |
| Energy contribution | -35.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.00 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.78 |
| SVM RNA-class probability | 0.999949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271516 117 - 23771897 GAAAAUGGGAAAUCCUUUGCUUUUAGCCU---CCUGUUCGCUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAA ......(((....)))(((((((((....---..(((..(((((.(((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))))) ( -36.90) >DroSec_CAF1 155032 117 - 1 GAAAAUGGGAAAUCCUUUGCUUUUAGCCU---CCUGUUCGCUUUUUGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAA ......(((....)))(((((((((....---..(((..((((((.((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))))) ( -34.80) >DroSim_CAF1 150065 117 - 1 GAAAAUGGGAAAUCCUUUGCUUUUAGCCU---CCUGUUCGCUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAA ......(((....)))(((((((((....---..(((..(((((.(((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))))) ( -36.90) >DroEre_CAF1 157875 117 - 1 GAAAAUGGGAAAUCCUUUGCUUUUAGCCU---CCUGUUCGCUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAA ......(((....)))(((((((((....---..(((..(((((.(((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))))) ( -36.90) >DroYak_CAF1 162680 120 - 1 GAAAAUGGGAAAUCCUUUGCUUUUGGCCUCCUCCUGUUCGCUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCCA ......(((....)))..(((((((.........(((..(((((.(((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))).. ( -34.70) >consensus GAAAAUGGGAAAUCCUUUGCUUUUAGCCU___CCUGUUCGCUUUUCGCUGUUUGCUCAGCGAAAGUACAAAUUGCAUGGCGCCCAGCUCAUUAAGAGUCGACAUGCAAUUAUAAAAGCAA ......(((....)))(((((((((.........(((..(((((.(((((......)))))))))))))(((((((((.((....((((.....)))))).))))))))).))))))))) (-35.94 = -35.82 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:30 2006