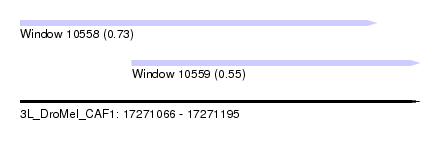
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,271,066 – 17,271,195 |
| Length | 129 |
| Max. P | 0.733903 |

| Location | 17,271,066 – 17,271,181 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 90.12 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -22.60 |
| Energy contribution | -23.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271066 115 + 23771897 GCUUUUCCACACACACCCACACACACACCCGAUGGAAAACACGGCAGAGAGUGAAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAGGACUGCA ((...(((........((((........(((.((.....)))))..(((.((....-..))))).(((((......)))))..)))).......((((.....)))).)))..)). ( -26.30) >DroSec_CAF1 154505 114 + 1 GCUUUUCCACACACACCCACACACACACACGUUGGAAAACACGGUGGAGAGUGAAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG-ACUGCA ((............................(((((....(((...((((.((....-..))))))(((((......)))))..))).)))))..((((.....))))..-...)). ( -28.30) >DroEre_CAF1 157329 112 + 1 GCUUUUCCACACACAC--ACACACACACCCGUUGGAAAACACGGCGGAGAGUGGAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAGCACUCACAG-UCUGCA ((..............--............(((((....(((...((((.((....-..))))))(((((......)))))..))).)))))..((((.....))))..-...)). ( -28.60) >DroYak_CAF1 162165 105 + 1 GCUUUUCCACACA----------CACACCCGCUGGAAAACACGGCGUAGAGUGAAAAAAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG-UGUGCA ...........((----------(((...(((((.......)))))..(((((............(((((......)))))..(((.....)))......)))))...)-)))).. ( -29.00) >consensus GCUUUUCCACACACAC__ACACACACACCCGUUGGAAAACACGGCGGAGAGUGAAA_AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG_ACUGCA ((............................(((((....(((...((((.((.......))))))(((((......)))))..))).)))))..((((.....))))......)). (-22.60 = -23.60 + 1.00)

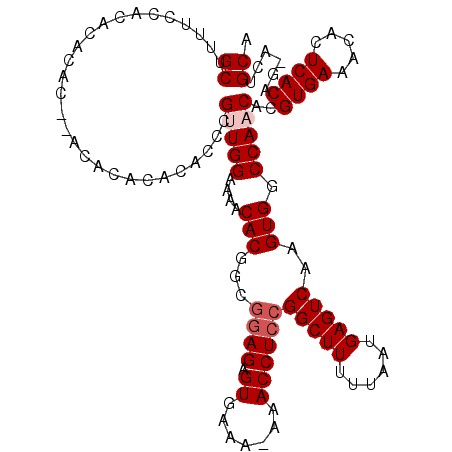

| Location | 17,271,102 – 17,271,195 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.26 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.44 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17271102 93 + 23771897 AAACACGGCAGAGAGUGAAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAGGACUGCACUGAGAACAUUCAU-------------------------- ......(((.(((.((....-..)))))..)))(((((.((((((..(((.....)))((((.....))))..)))).)).)))))........-------------------------- ( -20.10) >DroSec_CAF1 154541 118 + 1 AAACACGGUGGAGAGUGAAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG-ACUGCACUGAGAACUUUCAUAUGUAAUGAAGGUAUGAAAAAUAUCU ......(.(((((.((....-..))))))).).(((((.((((((..(((.....)))((((.....)))).)-))).)).)))))((((((((.....))))))))............. ( -30.50) >DroSim_CAF1 149571 118 + 1 AAACACGGCGGAGAGUGAAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG-AAUGCACUGAGAACAUUCAUAUGUAAUGAAGGUAUGAAUAAUAUCU ......(((((((.((....-..))))))(((((......))))).....))).(((..((((....((.(((-......))).))...))))..))).....((((((.....)))))) ( -29.00) >DroEre_CAF1 157363 109 + 1 AAACACGGCGGAGAGUGGAA-AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAGCACUCACAG-UCUGCACUGAAAACAUUAAUUUUUAAUGAAGA---------UGCAC ......(((((((.((....-..))))))(((((......))))).....))).....((((.....)))).(-(((.((.((((((......)))))).)).)))---------).... ( -27.10) >DroYak_CAF1 162191 110 + 1 AAACACGGCGUAGAGUGAAAAAAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG-UGUGCACUGAGAACAUUCAUUAUUAAUGAAGA---------UUUAU ..((((...((.(((((............(((((......)))))..(((.....)))......))))))).)-))).......(((..(((((.....)))))..---------))).. ( -22.10) >consensus AAACACGGCGGAGAGUGAAA_AAACCUCCGGCUUUUUAAUGAGUCAAGUGGCCAACACGUGAAACACUCACAG_ACUGCACUGAGAACAUUCAUAUGUAAUGAAGA_________UAUAU ......(((((((.((.......))))))(((((......))))).....))).....((((.....))))................................................. (-18.20 = -18.44 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:26 2006