
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,270,195 – 17,270,475 |
| Length | 280 |
| Max. P | 0.999265 |

| Location | 17,270,195 – 17,270,315 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -25.78 |
| Energy contribution | -26.78 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17270195 120 + 23771897 UCUUGGCUAAAAUAGUUUGCCAACAGUUGCUCCGCGCUGGCACAUUAACGUGGCCAAGGCGGCAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG ....(((..(((((.((((((...(((((((((((..((((.(((....)))))))..))))...............)))))))...))))))..........)))))..)))....... ( -31.96) >DroSec_CAF1 153657 120 + 1 UCUUGGCUAAAAUAGUUUGCCAACAGUUGCUCCUCGCUUGCACAUUAACGUGGCCAAGGCGGCAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG ....(((..(((((.((((((...(((((((.........(((......)))(((.....)))..............)))))))...))))))..........)))))..)))....... ( -25.50) >DroSim_CAF1 148683 120 + 1 UCUUGGCUAAAAUAGUUUGCCAACAGUUGCUCCUCGCUGGCACAUUAACGUGGCCAAGGCGGCAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG (((((((((...((((.(((((.(((......)).).))))).))))...))))))))).((((..............((........))(((((.........)))))))))....... ( -27.30) >DroEre_CAF1 156466 120 + 1 UCUUGGCCAAAAUAGUUUGCCAACAGUUGCUCCGCGCUGGCACAUUAACGUGGCCAAGGCGGCAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG ....(((..(((((.((((((...(((((((((((..((((.(((....)))))))..))))...............)))))))...))))))..........)))))..)))....... ( -31.06) >DroYak_CAF1 161280 119 + 1 UCUUGGCUAAAAUAGUUUGCGAACAGUUGCUCCGCGCUGGCACAUUAACGUGGCCAAGGCG-CAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG ....(((..(((((.((((((((.(((((((..((((((((.(((....))))))..))))-)...((......)).)))))))))).)))))..........)))))..)))....... ( -28.70) >consensus UCUUGGCUAAAAUAGUUUGCCAACAGUUGCUCCGCGCUGGCACAUUAACGUGGCCAAGGCGGCAAAUAAUUCAAUAAAGCAACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUG ....(((..(((((.((((((...(((((((((((..((((.(((....)))))))..))))...............)))))))...))))))..........)))))..)))....... (-25.78 = -26.78 + 1.00)

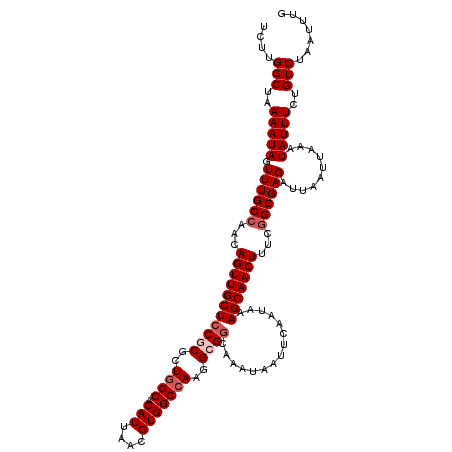

| Location | 17,270,195 – 17,270,315 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.96 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17270195 120 - 23771897 CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUGCCGCCUUGGCCACGUUAAUGUGCCAGCGCGGAGCAACUGUUGGCAAACUAUUUUAGCCAAGA ..(((((((.(....).)))))))......(((((.(((((((...............((((.(((((.(((....)))))))).))))))))))).))))).................. ( -33.06) >DroSec_CAF1 153657 120 - 1 CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUGCCGCCUUGGCCACGUUAAUGUGCAAGCGAGGAGCAACUGUUGGCAAACUAUUUUAGCCAAGA ..(((((((.(....).)))))))......(((((.(((((((((............(((.....)))..((((........)))).))))))))).))))).................. ( -27.60) >DroSim_CAF1 148683 120 - 1 CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUGCCGCCUUGGCCACGUUAAUGUGCCAGCGAGGAGCAACUGUUGGCAAACUAUUUUAGCCAAGA ..(((((((.(....).)))))))......(((((.(((((((((....((....))..(((..((((.(((....)))))))))).))))))))).))))).................. ( -28.90) >DroEre_CAF1 156466 120 - 1 CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUGCCGCCUUGGCCACGUUAAUGUGCCAGCGCGGAGCAACUGUUGGCAAACUAUUUUGGCCAAGA ..(((((((.(....).)))))))......(((((.(((((((...............((((.(((((.(((....)))))))).))))))))))).))))).................. ( -33.06) >DroYak_CAF1 161280 119 - 1 CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUG-CGCCUUGGCCACGUUAAUGUGCCAGCGCGGAGCAACUGUUCGCAAACUAUUUUAGCCAAGA ..(((((((.(....).)))))))......((.(((((((((((............((-(((..((((.(((....)))))))))))))))))))).))))).................. ( -27.50) >consensus CAAAUUAGACAGAAAUAUUUAAUUAAUCUCGCCGAAAGUUGCUUUAUUGAAUUAUUUGCCGCCUUGGCCACGUUAAUGUGCCAGCGCGGAGCAACUGUUGGCAAACUAUUUUAGCCAAGA ..(((((((.(....).)))))))......(((((.(((((((...............((((.(((((.(((....)))))))).))))))))))).))))).................. (-26.96 = -27.96 + 1.00)

| Location | 17,270,275 – 17,270,395 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -29.38 |
| Energy contribution | -29.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17270275 120 + 23771897 AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCAGGGGGUGCGAACUAGAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUA .(((..(((((((((.........))))).(((......)))(((.....)))...))).)..)))(((..((.....))..))).....(((((((.((((((...))))))))))))) ( -34.00) >DroSec_CAF1 153737 120 + 1 AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCAGGGGGAGCGAACUAAAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUA ..((..(((((((((.........))))).(((......)))(((.....)))...))).)..)).(((..((.....))..))).....(((((((.((((((...))))))))))))) ( -33.10) >DroSim_CAF1 148763 120 + 1 AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCAGGGGGAGCGAACUAAAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUA ..((..(((((((((.........))))).(((......)))(((.....)))...))).)..)).(((..((.....))..))).....(((((((.((((((...))))))))))))) ( -33.10) >DroEre_CAF1 156546 119 + 1 AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCA-GGGGAGCGAACUAAAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGCCAAACUUGCUUA ..(((..((((((((.........))))).(((......)))(((.....)))...))).-.))).(((..((.....))..))).....(((((((.(((((.....)))))))))))) ( -29.30) >DroYak_CAF1 161359 120 + 1 AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCACAGGGAGCGAACUAAAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGCCAAACUUGCUUA ...((((.(((((((.........))))).(((......)))))......((((.(.((((..((......))...)))).)))))))))(((((((.(((((.....)))))))))))) ( -28.80) >consensus AACUUUCGGCGAGAUUAAUUAAAUAUUUCUGUCUAAUUUGACGCAAUUUAUGCAAUGCCAGGGGGAGCGAACUAAAGUGGGGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUA ...((((((((((((.........))))).(((......)))(((.....)))...))).......(((..((.....))..))).))))(((((((.(((((.....)))))))))))) (-29.38 = -29.38 + 0.00)


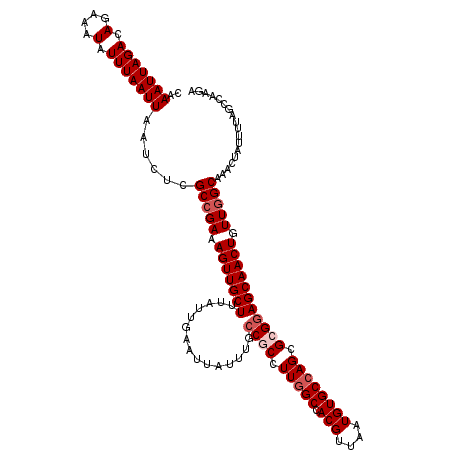
| Location | 17,270,355 – 17,270,475 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -34.89 |
| Consensus MFE | -32.26 |
| Energy contribution | -32.46 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17270355 120 + 23771897 GGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUAUCUAUCUGGUGAAACUUUUAUUAAUUACAAAGGAUACACAUCAACAUUAGCCCAAGUGAGCUGCGGAUGUGGCGUCACAU ((.((....((((((((.((((((...)))))))))))))).(((((.((((............))))...))))).............))))..((((((..(....)..)).)))).. ( -33.20) >DroSec_CAF1 153817 120 + 1 GGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUAUCUAUCUGGUGAAACUUUUAUUAAUUACAAAGGAUAUACAUCAACAUUAGUCCAAGUGAGCUGCGGAAGUGGCGUCACAU ((((((((.((((((((.((((((...))))))))))))))...))))......(((((.........))))).....)))).............((((((..(....)..)).)))).. ( -35.30) >DroSim_CAF1 148843 120 + 1 GGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUAUCUAUCUGGUGAAACUUUUAUUAAUUACAAAGGAUACACAUCAACAUUAGUCCAAGUGAGCUGCGGAAGUGGCGUCACAU ((((((((.((((((((.((((((...))))))))))))))...))))(((...((((((.....)).))))..))).)))).............((((((..(....)..)).)))).. ( -36.10) >DroEre_CAF1 156625 120 + 1 GGUGCAGAAAUGAGCAAAGUUUGCCGGCCAAACUUGCUUAUCUAUCUGGUGAAACUUUUAUUAAUUACAAAGGAUACACAUCAACAUUAGCCCAAGUGAGCUGCGGAAGUGGCUUCACAU ((.((....((((((((.(((((.....))))))))))))).(((((.((((............))))...))))).............))))..((((((..(....)..)).)))).. ( -34.70) >DroYak_CAF1 161439 120 + 1 GGUGCAGAAAUGAGCAAAGUUUGCCGGCCAAACUUGCUUAGCUAUCUGCUGAAACUUUUAUUAAUUACAAAGGAUACACAUCAACAUUAGUCCAAGUGAGCUGCGGAAGUGGCUUCACAU ...(((((..(((((((.(((((.....))))))))))))....)))))......................((((..............))))..((((((..(....)..)).)))).. ( -35.14) >consensus GGUGCAGAAAUGAGCAAAGUUUGCCGGGCAAACUUGCUUAUCUAUCUGGUGAAACUUUUAUUAAUUACAAAGGAUACACAUCAACAUUAGUCCAAGUGAGCUGCGGAAGUGGCGUCACAU ((((((((.((((((((.(((((.....)))))))))))))...))))......(((((.........))))).....)))).............((((((..(....)..)).)))).. (-32.26 = -32.46 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:23 2006