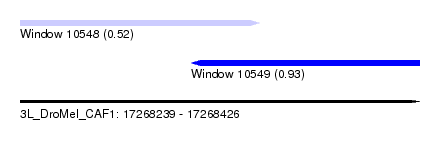
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,268,239 – 17,268,426 |
| Length | 187 |
| Max. P | 0.932934 |

| Location | 17,268,239 – 17,268,351 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.23 |
| Mean single sequence MFE | -41.34 |
| Consensus MFE | -31.32 |
| Energy contribution | -31.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17268239 112 + 23771897 UUCCCUUGCAAUUUUUGUACGCUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGUGUGGCUGAUGGCCGCAUUGAAUGGCCAU--------GGGGCGUGGCACCGCCCCUUGUGCCAUUA ......((((.....)))).((....))....(((((.........)))))(.(((((((.....))))))).)((((((((.--------((((((......)))))).)).)))))). ( -39.30) >DroSec_CAF1 151662 112 + 1 UUCCCUUGCAAUUUUUGUACGCUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGUGUGGCUGAUGGCGGCAUUGAAUGGCCAG--------GGGGCGUGGCACCGCACCAUGUGCCAUUA .(((((.((.((((.(((.(((((..(((.....(((((((((...))))).)))).)))..).)))))))..)))).)).))--------)))..(((((((........))))))).. ( -38.70) >DroSim_CAF1 146713 112 + 1 UUCCCUUGCAAUUUUUGUACGUUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGUGUGGCUGAUGGCGGCAUUGAAUGGCCAG--------GGGGCGUGGCACCGCCCCAUGUGCCAUUA ...............(((.(((((..(((.....(((((((((...))))).)))).)))..).)))))))...((((((((.--------((((((......)))))).)).)))))). ( -37.60) >DroEre_CAF1 154657 111 + 1 AUCCCCUGCAAUUUUUGUACGCUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGUGUGGCUGAUGGCGGCAU-GAAUGGGCAG--------GGGGCGUGGCACCGCCCCACGUGCCAUUA .((((((((.((((.(((.(((((..(((.....(((((((((...))))).)))).)))..).))))))).-))))..))))--------)))).(((((((........))))))).. ( -46.00) >DroYak_CAF1 159319 120 + 1 AUCCCUUGCAAUUUUUGUACGCUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGGGUGGCUGAUGGCGGCAUUGAAUGGCCAGGAGGCGAGGGGGCGUGGCACCGCCCCAUGUGCCAUUA ...(((.((.((((.(((.(((((..(((...(((((..((((...)))))).))).)))..).)))))))..)))).)).))).((((..((((((......))))))...)))).... ( -45.10) >consensus UUCCCUUGCAAUUUUUGUACGCUUUGGCUUUUCUCGCAUUCAUUACGUGAGCUGUGUGGCUGAUGGCGGCAUUGAAUGGCCAG________GGGGCGUGGCACCGCCCCAUGUGCCAUUA ................(((((...(((((.(((..((........(((.((((....)))).)))...))...))).))))).........((((((......)))))).)))))..... (-31.32 = -31.56 + 0.24)


| Location | 17,268,319 – 17,268,426 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.72 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -35.58 |
| Energy contribution | -36.26 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17268319 107 - 23771897 AAAACAACGAGGACUAAAUGAAAUGCUCCUUG-----GAGUCGGGGAUGUGCCACCGCAGAAGGGGAGCUCAUGCUGGGCUAAUGGCACAAGGGGCGGUGCCACGCCCC--------AUG ...((..(((((((..........).))))))-----..))......(((((((((.(....).))(((((.....)))))..))))))).((((((......))))))--------... ( -41.30) >DroSec_CAF1 151742 107 - 1 AAAACAACGAGGGCUAAAUGAAAUGCUCCUUG-----GAGUCGGGUAUGUGCCACCGCAGAAGGGGAGCUCAUGCUGGGCUAAUGGCACAUGGUGCGGUGCCACGCCCC--------CUG ...((..(((((((..........)).)))))-----..)).(((.((((((((((.(....).))(((((.....)))))..))))))))((((.(....).))))))--------).. ( -39.80) >DroSim_CAF1 146793 107 - 1 AAAACAACGAGGGCUAAAUGAAAUGCUCCUUG-----GAGUCGGGUAUGUGCCACCGCAGAAGGGGAGCUCAUGCUGGGCUAAUGGCACAUGGGGCGGUGCCACGCCCC--------CUG ...((..(((((((..........)).)))))-----..))(((((((((((((((.(....).))(((((.....)))))..)))))))))(((((......))))))--------))) ( -44.20) >DroEre_CAF1 154736 105 - 1 -AAACAACGAGGGCUAAAUGAAAUGCUCCUUG-----GAGUCGGGCAUGUGCCACCACAGAAG-GGAGCUCAUGCUGGGCUAAUGGCACGUGGGGCGGUGCCACGCCCC--------CUG -..((..(((((((..........)).)))))-----..))(((((((((((((((......)-).(((((.....)))))..)))))))))(((((......))))))--------))) ( -44.40) >DroYak_CAF1 159399 119 - 1 -AAACAACGAGGGCUAAAUGAAAUGCUCCUUGGCUUCGAGUCGGGGAAGUGCCACCACAGACGGGGAGCUCAUGCUGGGCUAAUGGCACAUGGGGCGGUGCCACGCCCCCUCGCCUCCUG -......(((((((..........)).))))).....(((.((((...((((((((......))..(((((.....)))))..))))))..((((((......)))))))))).)))... ( -47.80) >consensus AAAACAACGAGGGCUAAAUGAAAUGCUCCUUG_____GAGUCGGGGAUGUGCCACCGCAGAAGGGGAGCUCAUGCUGGGCUAAUGGCACAUGGGGCGGUGCCACGCCCC________CUG .......(((((((..........)).)))))..............((((((((((.(....).))(((((.....)))))..))))))))((((((......))))))........... (-35.58 = -36.26 + 0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:16 2006