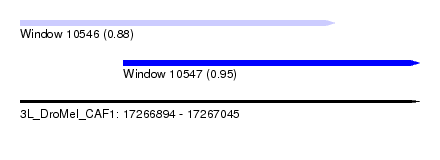
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,266,894 – 17,267,045 |
| Length | 151 |
| Max. P | 0.946820 |

| Location | 17,266,894 – 17,267,013 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -34.52 |
| Energy contribution | -34.76 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17266894 119 + 23771897 UC-AAAAGGGACGACACGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAU ..-..................((....((..((((((((..(((((..(((((....(((((.....)))))...))))).)))))))))))))..))....)).(((.....))).... ( -36.60) >DroSec_CAF1 150349 120 + 1 UCCAAAAGGGACGACACGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAU (((.....)))..........((....((..((((((((..(((((..(((((....(((((.....)))))...))))).)))))))))))))..))....)).(((.....))).... ( -38.10) >DroSim_CAF1 145401 120 + 1 UCCAAAAGGGACGACCCGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGAAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAU .......(((....(((....((....((..((((((((..(((((..(((((....(((((.....)))))...))))).)))))))))))))..))....)).))).....))).... ( -41.60) >DroEre_CAF1 153382 119 + 1 UCCAGGA-GGACGACACGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUCCAGGGACUUACCCAAAU (((....-)))........(((..(((((..((((((((..(((((..(((((....(((((.....)))))...))))).))))))))))))).((.....))...)))))..)))... ( -39.10) >DroYak_CAF1 157991 118 + 1 UCCAAAA-GGACGACACGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUCGCAG-GACUUACCCAAAU (((....-)))........(((..(((((..((((((((..(((((..(((((....(((((.....)))))...))))).)))))))))))))..(....)....-)))))..)))... ( -35.60) >consensus UCCAAAAGGGACGACACGAUGGCCAGGUCAGUUCGCUGUGAUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAU (((.........(((.(........))))..((((((((..(((((..(((((....(((((.....)))))...))))).))))))))))))).))).......(((.....))).... (-34.52 = -34.76 + 0.24)


| Location | 17,266,933 – 17,267,045 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.70 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -35.54 |
| Energy contribution | -35.42 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.946820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17266933 112 + 23771897 AUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAUGGCAAG--------UGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))((((((((.....))))).(..((...((((.(((((...--------.)))))...))))...))..).))). ( -37.90) >DroSec_CAF1 150389 112 + 1 AUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAUGGCAAG--------UGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))((((((((.....))))).(..((...((((.(((((...--------.)))))...))))...))..).))). ( -37.90) >DroSim_CAF1 145441 112 + 1 AUUGGCAGAAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAUGGCAAG--------UGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))((((((((.....))))).(..((...((((.(((((...--------.)))))...))))...))..).))). ( -37.50) >DroEre_CAF1 153421 120 + 1 AUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUCCAGGGACUUACCCAAAUGGCAAUUGUCGCGGGGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))(((((....((.....)).(((.....))).....))...((((((((..(((....)))..))))))))))). ( -39.90) >DroYak_CAF1 158030 111 + 1 AUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUCGCAG-GACUUACCCAAAUGGCAAC--------GGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))((((((((.....)))))..-(((...((((.(((((...--------.)))))...)))))))......))). ( -39.00) >consensus AUUGGCAGGAGUCAUCAGGCAAGUUAAUUGCCCACGAUUUAGCCAAGCGGCGAAAGGAAUUUGCAGGGACUUACCCAAAUGGCAAG________UGUCGUGAGUGGGGUCGUGACACGCG .(((((..(((((....(((((.....)))))...))))).)))))((((((((.....)))))...(((...((((.(((((............)))))...)))))))......))). (-35.54 = -35.42 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:14 2006