
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,265,833 – 17,266,064 |
| Length | 231 |
| Max. P | 0.997733 |

| Location | 17,265,833 – 17,265,931 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -21.42 |
| Energy contribution | -21.62 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265833 98 + 23771897 AAUACU------------------CAUAAAGUAUGCAAAACUUAUGCACCAUCCAAGCGC---CGCCCUUUUC-GGGGGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUG ......------------------((((((..((((...((((((...(((....(((..---((((((....-.))))))..)))..)))....)))))).))))))))))........ ( -31.50) >DroSec_CAF1 149331 101 + 1 AAUACUC--------------ACUCAUAAAGUAUGCAAAACUUAUGCACCAUCCAAGCGC---CGCACCUUUC-GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUG .......--------------...((((((..((((...((((((......(((((..((---(((.(((...-.)-)).)))))..)))))...)))))).))))))))))........ ( -30.40) >DroSim_CAF1 144361 101 + 1 AAUACUC--------------GCUCAUAAAGUAUGCAAAACUUAUGCACCAUCCAAGCGC---CGCCCCUUUC-GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUG .......--------------(..((((((..((((...((((((...(((....(((..---((((((....-))-))))..)))..)))....)))))).))))))))))..)..... ( -34.50) >DroEre_CAF1 152382 96 + 1 AA------------------UACUCAUAAAGUAUGCAAAACUUAUGCACCAUCCUCGCAC---CGCCCUUUU--GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUG ..------------------....((((((..((((...((((((...(((.....((..---(((((....--.)-))))..))...)))....)))))).))))))))))........ ( -28.10) >DroYak_CAF1 156849 119 + 1 AACACUCAUACUCAUACUCAUACUCAUAAAGUAUGCAAAACUUAUGCACCAUCCCACCACCAGCGCCCUUUUUGGG-GGCGUGGCUGUUGGACAAGUAAGUGGUAUUUUAUGUUCAUUUG ......((((...(((((((((((.....))))))....((((((......(((.((..(((.((((((.....))-)))))))..)).)))...)))))))))))..))))........ ( -30.40) >consensus AAUACUC______________ACUCAUAAAGUAUGCAAAACUUAUGCACCAUCCAAGCGC___CGCCCUUUUC_GG_GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUG ........................((((((..((((...((((((...(((....(((.(...((((((.....)).)))).))))..)))....)))))).))))))))))........ (-21.42 = -21.62 + 0.20)



| Location | 17,265,833 – 17,265,931 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.29 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -18.82 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265833 98 - 23771897 CAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCACGCCCCC-GAAAAGGGCG---GCGCUUGGAUGGUGCAUAAGUUUUGCAUACUUUAUG------------------AGUAUU ..((((..((((((((((.....((((((.((.((.((((((((((-.....))).)---)))..))).)).)).))))))...))))..))))))------------------..)))) ( -30.10) >DroSec_CAF1 149331 101 - 1 CAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCACGCC-CC-GAAAGGUGCG---GCGCUUGGAUGGUGCAUAAGUUUUGCAUACUUUAUGAGU--------------GAGUAUU .............(((((((((((...(((((..(((..(((.-((-....)).)))---..)))))))).(((((.......)))))......)))))--------------).))))) ( -32.80) >DroSim_CAF1 144361 101 - 1 CAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCACGCC-CC-GAAAGGGGCG---GCGCUUGGAUGGUGCAUAAGUUUUGCAUACUUUAUGAGC--------------GAGUAUU ....((..((((((((((.(((((.(..((((..(((..((((-((-....))))))---..)))))))..)....)))))...))))..))))))..)--------------)...... ( -36.00) >DroEre_CAF1 152382 96 - 1 CAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCACGCC-CC--AAAAGGGCG---GUGCGAGGAUGGUGCAUAAGUUUUGCAUACUUUAUGAGUA------------------UU ..((((..((((((((((.(((((..((((((.(.((..((((-(.--....)))))---..))...).))).))))))))...))))..))))))..))------------------)) ( -27.20) >DroYak_CAF1 156849 119 - 1 CAAAUGAACAUAAAAUACCACUUACUUGUCCAACAGCCACGCC-CCCAAAAAGGGCGCUGGUGGUGGGAUGGUGCAUAAGUUUUGCAUACUUUAUGAGUAUGAGUAUGAGUAUGAGUGUU ........((((..((((((..(((((((((.((.((((((((-(.......))))).)))).)).)))).(((((.......))))).......))))))).))))...))))...... ( -35.80) >consensus CAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCACGCC_CC_GAAAAGGGCG___GCGCUUGGAUGGUGCAUAAGUUUUGCAUACUUUAUGAGU______________GAGUAUU ........((((((((((.(((((.(..((((...((..((((.(.......)))))...))...))))..)....)))))...))))..))))))........................ (-18.82 = -19.38 + 0.56)


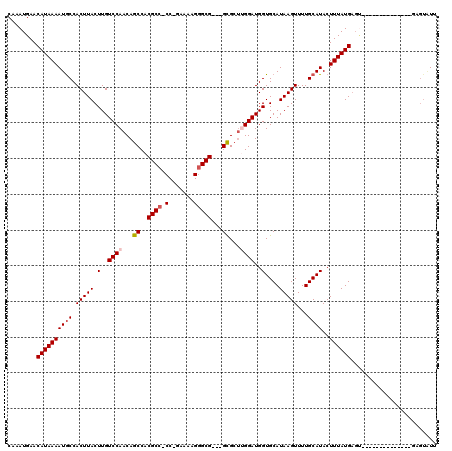
| Location | 17,265,855 – 17,265,971 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -27.84 |
| Energy contribution | -28.00 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265855 116 + 23771897 CUUAUGCACCAUCCAAGCGC---CGCCCUUUUC-GGGGGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACUCCUCUUCUUCAAUGACAAAAUGGCAUUGU ...((((........(((((---((((((....-.)))))(((((...((((((..((((......))))))))))...))))))))).)).....((......)).......))))... ( -35.10) >DroSec_CAF1 149357 115 + 1 CUUAUGCACCAUCCAAGCGC---CGCACCUUUC-GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACUCCUCUUCUUCAAUGCCAAAAUGGCAUUGU ....(((....(((((..((---(((.(((...-.)-)).)))))..))))).......((((((.............)))))).)))............(((((((.....))))))). ( -39.62) >DroSim_CAF1 144387 115 + 1 CUUAUGCACCAUCCAAGCGC---CGCCCCUUUC-GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACCUCUCUUCUUCAAUGCCAAAAUGGCAUUGU .................(((---((((((....-))-)))(((((...((((((..((((......))))))))))...)))))))))............(((((((.....))))))). ( -43.90) >DroEre_CAF1 152404 112 + 1 CUUAUGCACCAUCCUCGCAC---CGCCCUUUU--GG-GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGACU--CCCUCUCCUUCAAUGCCAAAAUGGCAUUGU ..............((((..---(((((....--.)-)))).(((...((((((..((((......))))))))))...)))))))..--..........(((((((.....))))))). ( -36.00) >DroYak_CAF1 156889 117 + 1 CUUAUGCACCAUCCCACCACCAGCGCCCUUUUUGGG-GGCGUGGCUGUUGGACAAGUAAGUGGUAUUUUAUGUUCAUUUGCCGCGACU--CGCUCUUCCUCAAUGCCAAAAUGGCAUUGU .....((......(((.(((((.((((((.....))-))))))).)).)))........((((((.............))))))....--.)).......(((((((.....))))))). ( -38.22) >consensus CUUAUGCACCAUCCAAGCGC___CGCCCUUUUC_GG_GGCGUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACCCCUCUUCUUCAAUGCCAAAAUGGCAUUGU ........................(((((.....)).)))(((((...((((((..((((......))))))))))...)))))................(((((((.....))))))). (-27.84 = -28.00 + 0.16)

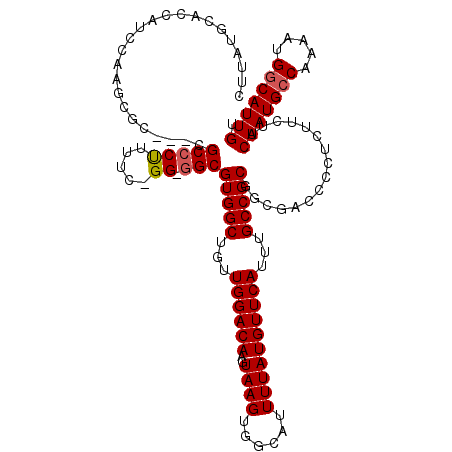

| Location | 17,265,891 – 17,266,011 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -34.46 |
| Energy contribution | -33.82 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265891 120 + 23771897 GUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACUCCUCUUCUUCAAUGACAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAA ((.(((............))).)).((((((((.(((((..((((((((...........(((((.(.....).)))))............)))))...))).))))).))))))))... ( -29.10) >DroSec_CAF1 149392 120 + 1 GUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACUCCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAA ((.(((............))).)).((((((((.(((((..((((((((...........(((((((.....)))))))............)))))...))).))))).))))))))... ( -36.20) >DroSim_CAF1 144422 120 + 1 GUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACCUCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAA ((.(((............))).)).((((((((.(((((..((((((((...........(((((((.....)))))))............)))))...))).))))).))))))))... ( -36.20) >DroEre_CAF1 152438 118 + 1 GUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGACU--CCCUCUCCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGGUGGGG (((((...((((((..((((......))))))))))...)))))....--.((((.((...((((((.....))))))((((.....((((((((....))))))))))))..)).)))) ( -40.90) >DroYak_CAF1 156928 118 + 1 GUGGCUGUUGGACAAGUAAGUGGUAUUUUAUGUUCAUUUGCCGCGACU--CGCUCUUCCUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGACGGCG ...(((((((((..(((..((((((.............))))))....--.)))..)))..((((((.....))))))((((.....((((((((....))))))))))))..)))))). ( -37.32) >consensus GUGGCUGUUGGACAAGUAAGUGGCAUUUUAUGUUCAUUUGCCGCGGCGACCCCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAA (((((...((((((..((((......))))))))))...)))))...........((((((((((((.....)))))(((((.....((((((((....)))))))))))))))).)))) (-34.46 = -33.82 + -0.64)

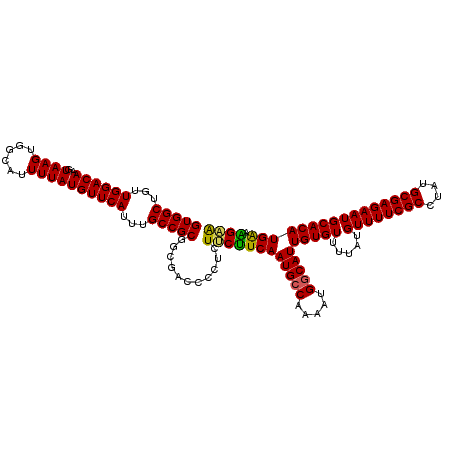
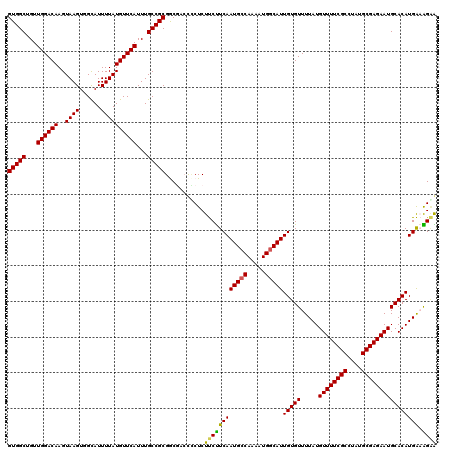
| Location | 17,265,891 – 17,266,011 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -24.95 |
| Energy contribution | -23.95 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.914002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265891 120 - 23771897 UUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGUCAUUGAAGAAGAGGAGUCGCCGCGGCAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCAC ........(((((......)))))(((..............(((((.(.....).))))).....((.((((....(.((((.............)))).)..)))).)).....))).. ( -22.52) >DroSec_CAF1 149392 120 - 1 UUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGGAGUCGCCGCGGCAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCAC ........(((((......)))))(((..............(((((((.....))))))).....((.((((....(.((((.............)))).)..)))).)).....))).. ( -29.62) >DroSim_CAF1 144422 120 - 1 UUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGAGGUCGCCGCGGCAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCAC ........(((((......)))))(((((............(((((((.....)))))))...........)))))..(((...((.(((................))).))...))).. ( -28.99) >DroEre_CAF1 152438 118 - 1 CCCCACCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGGAGAGGG--AGUCGCGGCAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCAC ........(((((......)))))(((..............(((((((.....)))))))..(((.((((--(((...((((.............)))))))).))).)))....))).. ( -33.82) >DroYak_CAF1 156928 118 - 1 CGCCGUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAGGAAGAGCG--AGUCGCGGCAAAUGAACAUAAAAUACCACUUACUUGUCCAACAGCCAC .(((((.((.(((.(((((((....))))............(((((((.....)))))))..)))..)))--.)).)))))....................................... ( -29.60) >consensus UUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGGGGUCGCCGCGGCAAAUGAACAUAAAAUGCCACUUACUUGUCCAACAGCCAC (((((((((((......((((....))))..))))......(((((((.....)))))))..)))).)))........(((...((.(((................))).))...))).. (-24.95 = -23.95 + -1.00)

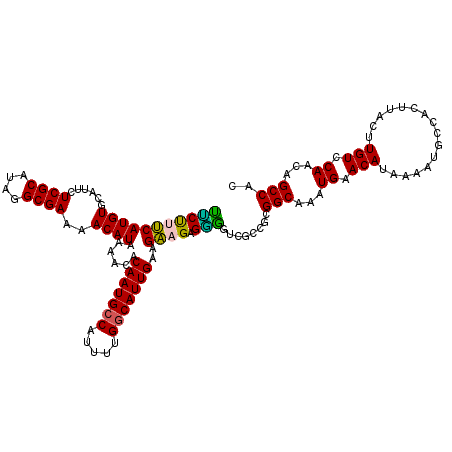

| Location | 17,265,931 – 17,266,032 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -21.04 |
| Energy contribution | -20.80 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265931 101 + 23771897 CCGCGGCGACUCCUCUUCUUCAAUGACAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAAGUG-------------------UGGGUACAGAUGGCAAUA ..((.((.(((((.(((((((((((.(.....).)))(((((.....((((((((....))))))))))))))).)))))).)-------------------.))))...).).)).... ( -25.30) >DroSec_CAF1 149432 98 + 1 CCGCGGCGACUCCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAAGUA-------------------UGGGUGCAGAGUGCA--- .(((.((.((((..(((((((((((((.....)))))(((((.....((((((((....))))))))))))))).))))))..-------------------.))))))...)))..--- ( -32.90) >DroSim_CAF1 144462 98 + 1 CCGCGGCGACCUCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAAGUA-------------------UGGGUGCAGAGUGCA--- .(((.((.((((..(((((((((((((.....)))))(((((.....((((((((....))))))))))))))).))))))..-------------------.))))))...)))..--- ( -34.00) >DroEre_CAF1 152478 115 + 1 CCGCGACU--CCCUCUCCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGGUGGGGAUGUGGAUGGUGGAUGUGGGUGUGGGUGCGGAGGGCA--- (((((((.--(((.((((..(((((((.....)))))))...((((((((..((((..(((......)))...))))..))))))))....))).).))).))...)))))......--- ( -43.70) >DroYak_CAF1 156968 99 + 1 CCGCGACU--CGCUCUUCCUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGACGGCGAUG-------------UUGGUGUGGGUGUGGAA---A--- ((((.(((--(((.((..(((((((((.....))))))((((.....((((((((....))))))))))))........)).)-------------..)).))))))))))..---.--- ( -35.40) >consensus CCGCGGCGACCCCUCUUCUUCAAUGCCAAAAUGGCAUUGUGUUUUAUGUUUUCGCCUAUGCGAGAAUGCACAUGAAAGAAGUG___________________UGGGUGCAGAGGGCA___ ..............(((((((((((((.....)))))(((((.....((((((((....)))))))))))))))).)))))....................................... (-21.04 = -20.80 + -0.24)



| Location | 17,265,931 – 17,266,032 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.87 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -21.82 |
| Energy contribution | -22.38 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265931 101 - 23771897 UAUUGCCAUCUGUACCCA-------------------CACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGUCAUUGAAGAAGAGGAGUCGCCGCGG .........((((..(.(-------------------(.(((((((..(((((......)))))(((((((.(((........)))...)))))))......))))))).)).)..)))) ( -19.30) >DroSec_CAF1 149432 98 - 1 ---UGCACUCUGCACCCA-------------------UACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGGAGUCGCCGCGG ---......((((.....-------------------...........(((((......)))))(((((............(((((((.....)))))))...........))))))))) ( -28.00) >DroSim_CAF1 144462 98 - 1 ---UGCACUCUGCACCCA-------------------UACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGAGGUCGCCGCGG ---......((((.....-------------------...........(((((......)))))(((((............(((((((.....)))))))...........))))))))) ( -28.00) >DroEre_CAF1 152478 115 - 1 ---UGCCCUCCGCACCCACACCCACAUCCACCAUCCACAUCCCCACCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGGAGAGGG--AGUCGCGG ---......((((....((.(((...(((..............(.((.(((((......))))))).).............(((((((.....)))))))..)))..)))--.)).)))) ( -32.80) >DroYak_CAF1 156968 99 - 1 ---U---UUCCACACCCACACCAA-------------CAUCGCCGUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAGGAAGAGCG--AGUCGCGG ---(---((((.............-------------..(((((...(((((......))))).)))))............(((((((.....))))))).))))).(((--...))).. ( -29.50) >consensus ___UGCACUCUGCACCCA___________________CACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAACACAAUGCCAUUUUGGCAUUGAAGAAGAGGGGUCGCCGCGG .........((((...................................(((((......)))))(((((............(((((((.....)))))))...........))))))))) (-21.82 = -22.38 + 0.56)



| Location | 17,265,971 – 17,266,064 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -13.70 |
| Consensus MFE | -9.83 |
| Energy contribution | -10.50 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
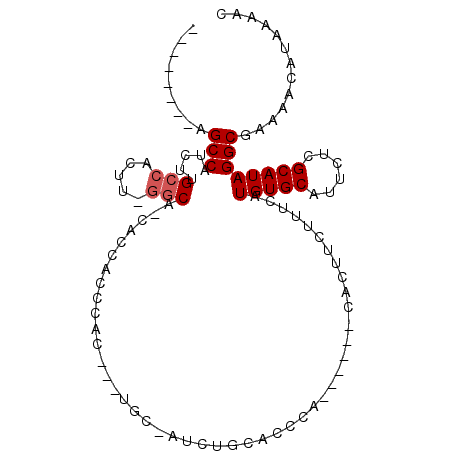
>3L_DroMel_CAF1 17265971 93 - 23771897 --------AGCCAUCUUGCCACUUUGCCACCACCACCCACUAUUGCCAUCUGUACCCA------CACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAAC --------..............((((((......................(((....)------)).........(((((......))))))))))).......... ( -11.70) >DroSim_CAF1 144502 96 - 1 ACCAUUACAGCCAUCUUGCCACAU-GGCA-CACCCCCCAC---UGCACUCUGCACCCA------UACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAAC .........(((....((((....-))))-..........---(((.....)))....------...........(((((......))))))))............. ( -17.10) >DroYak_CAF1 157006 90 - 1 ---------GCCAUCUUGCCACUU-UGCC-AUCUUGCCAC---U---UUCCACACCCACACCAACAUCGCCGUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAAC ---------((......((.....-.)).-.....))...---.---...................(((((...(((((......))))).)))))........... ( -12.30) >consensus ________AGCCAUCUUGCCACUU_GGCA_CACCACCCAC___UGC_AUCUGCACCCA______CACUUCUUUCAUGUGCAUUCUCGCAUAGGCGAAAACAUAAAAC .........(((.....(((.....)))...............................................(((((......))))))))............. ( -9.83 = -10.50 + 0.67)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:12 2006