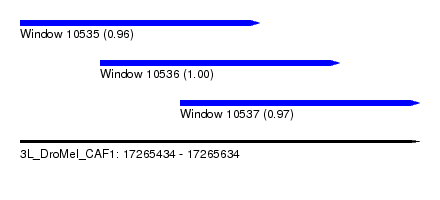
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,265,434 – 17,265,634 |
| Length | 200 |
| Max. P | 0.999959 |

| Location | 17,265,434 – 17,265,554 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -34.80 |
| Energy contribution | -34.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265434 120 + 23771897 GCGAAAAUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((((....((.((.(((....))).))))))))(((..((..((((((((....))))))))..))............((((((......))))))....((((((....)))))).))) ( -34.80) >DroSec_CAF1 148942 120 + 1 GCGAAACUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((....(((((((((((....)))..(((((((......))))......((((((((....(((......)))...))))))))))).))))))))....((((((....)))))).)). ( -37.70) >DroSim_CAF1 143975 120 + 1 GCGAAAAUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((((....((.((.(((....))).))))))))(((..((..((((((((....))))))))..))............((((((......))))))....((((((....)))))).))) ( -34.80) >DroEre_CAF1 152004 120 + 1 GCGAAAAUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((((....((.((.(((....))).))))))))(((..((..((((((((....))))))))..))............((((((......))))))....((((((....)))))).))) ( -34.80) >DroYak_CAF1 156487 120 + 1 GCGAAAAUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((((....((.((.(((....))).))))))))(((..((..((((((((....))))))))..))............((((((......))))))....((((((....)))))).))) ( -34.80) >consensus GCGAAAAUGUCGUAGCUUUUCAGCAACACUCGCAGCAAAGUGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCU ((((....((.((.(((....))).))))))))(((..((..((((((((....))))))))..))............((((((......))))))....((((((....)))))).))) (-34.80 = -34.80 + 0.00)

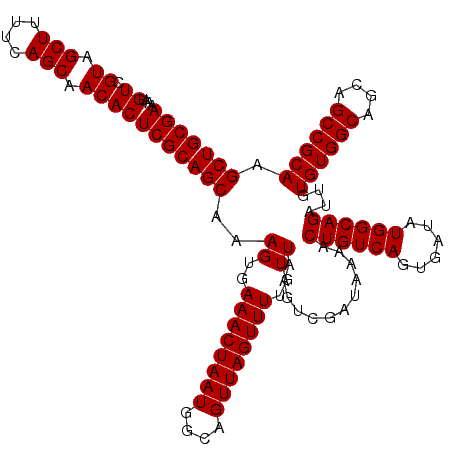
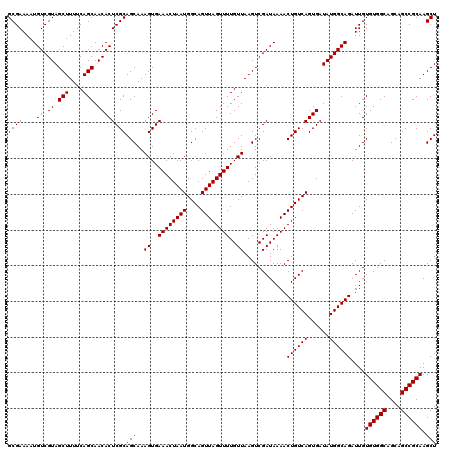
| Location | 17,265,474 – 17,265,594 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.54 |
| Mean single sequence MFE | -33.68 |
| Consensus MFE | -33.84 |
| Energy contribution | -33.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.89 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265474 120 + 23771897 UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUUUAAC .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))............ ( -33.30) >DroSec_CAF1 148982 116 + 1 UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU---- .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))........---- ( -33.30) >DroSim_CAF1 144015 116 + 1 UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU---- .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))........---- ( -33.30) >DroEre_CAF1 152044 119 + 1 UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCCAUUCUUUUCCCCACAUUUC-C .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))..........-. ( -35.20) >DroYak_CAF1 156527 116 + 1 UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUCCACUUU---- .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))........---- ( -33.30) >consensus UGAAACUAAUGGCAGUUAGUUUUGUUAAGUCGAUAAAACUGUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU____ .((((..(((((..(.((((((((((.....)))))))))).)(((((...((((((.((((((((....))))))....)).)))))).)))))..)))))..))))............ (-33.84 = -33.68 + -0.16)

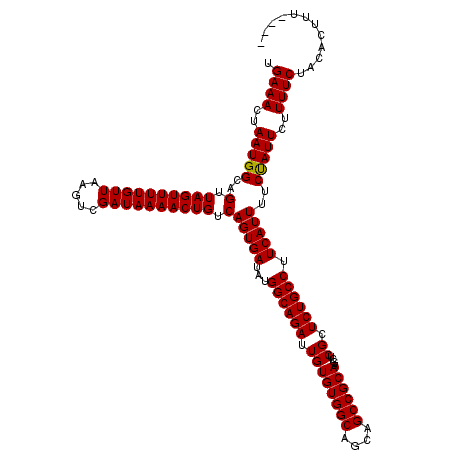
| Location | 17,265,514 – 17,265,634 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -22.52 |
| Consensus MFE | -22.02 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971895 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265514 120 + 23771897 GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUUUAACCGCUUUUUUUUCACUCCUGCCUGCUCCCCUUAACCCUCAC (..(((((...((((((.((((((((....))))))....)).)))))).)))))..).............................................................. ( -22.10) >DroSec_CAF1 149022 109 + 1 GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU-----------UUUUCACUCCUCCCUGCCCCCCUCCCCCCACAA (..(((((...((((((.((((((((....))))))....)).)))))).)))))..)..................-----------................................. ( -22.10) >DroSim_CAF1 144055 108 + 1 GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU------------UUUCACUCCUGCCUGCCCUCCUGCCCCCUCAC ....((((...((((((..(((.(((((.((((....)...))))))))..)))..))).................------------..........(....)......)))...)))) ( -23.50) >DroEre_CAF1 152084 112 + 1 GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCCAUUCUUUUCCCCACAUUUC-CCA-CUUUUUUCCACUCCUGGCUGCCUCCUUCC------CC ...........(((((...(((.(((((.((((....)...))))))))..)))........................-...-.......(((....)))))))).......------.. ( -22.80) >DroYak_CAF1 156567 90 + 1 GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUCCACUUU--------UUUUUGCCACUC----------------------CC (..(((((...((((((.((((((((....))))))....)).)))))).)))))..)..................--------............----------------------.. ( -22.10) >consensus GUCAGUGAUAUGGCAGAUUGUGUGGCAGCAGCCGCAAGCUCGCUCUGCCUUCAUUUUCUAUUCUUUUCUACACUUU________UUUUUUUCACUCCUGCCUGCCCCCCUCC_CCC_CAC (..(((((...((((((.((((((((....))))))....)).)))))).)))))..).............................................................. (-22.02 = -22.02 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:05 2006