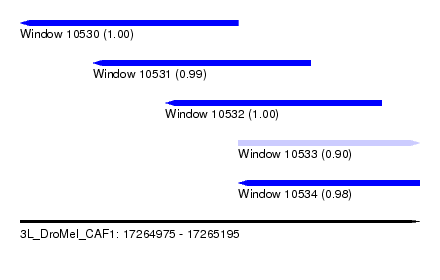
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,264,975 – 17,265,195 |
| Length | 220 |
| Max. P | 0.999333 |

| Location | 17,264,975 – 17,265,095 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.58 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -28.88 |
| Energy contribution | -28.84 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17264975 120 - 23771897 AACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...(((((((.((((((.......))))))..(.......)...)))))))(((.((((((..(..(((((..........)))))..).(....))))))))))............... ( -31.90) >DroSec_CAF1 148489 120 - 1 AACCGAACGAAUGCCUCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...(((((((..((..........))(((....)))........)))))))(((.((((((..(..(((((..........)))))..).(....))))))))))............... ( -27.30) >DroSim_CAF1 143523 120 - 1 AACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...(((((((.((((((.......))))))..(.......)...)))))))(((.((((((..(..(((((..........)))))..).(....))))))))))............... ( -31.90) >DroEre_CAF1 151542 119 - 1 AACCGAACGAAUGCCG-CCCAAUAGCGGCAAAGGCGAAAACAAAUCGUUUGUGCCCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...........(((((-((.....(.((((.((((((.......)))))).)))))(((.((.(..(((((..........)))))..).))...))))))))))............... ( -34.00) >DroYak_CAF1 156007 120 - 1 AACCGAACGAAUGUCGUCCCAAUAGCGGCGAAGGCGAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...........(((((((.(....(((((((..(((.((((.....)))).))))))))))..(..(((((..........)))))..).......).)))))))............... ( -28.90) >consensus AACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUUAAAAUGAACAUGAAAAGCGGCGGCAGAUAAAGAGCAAAUG ...(((((((.((((((.......))))))..(.......)...)))))))(((.((((((..(..(((((..........)))))..).(....))))))))))............... (-28.88 = -28.84 + -0.04)


| Location | 17,265,015 – 17,265,135 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.25 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -29.28 |
| Energy contribution | -29.44 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265015 120 - 23771897 GUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUU (((......................((....))(((((.....).......((((((.......))))))..))))....(((((.(((..(((.....)))..)))))))))))..... ( -34.20) >DroSec_CAF1 148529 120 - 1 GUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCUCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUU (((((...................(((....))).((......(((((((..((..........))(((....)))........)))))))(((.....))).)).....)))))..... ( -29.00) >DroSim_CAF1 143563 120 - 1 GUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUU (((......................((....))(((((.....).......((((((.......))))))..))))....(((((.(((..(((.....)))..)))))))))))..... ( -34.20) >DroEre_CAF1 151582 119 - 1 GUCUAUUUCAUUAAUACAAAUAAAGCCGGAAGGGGCCGAAAACCGAACGAAUGCCG-CCCAAUAGCGGCAAAGGCGAAAACAAAUCGUUUGUGCCCGCUGCAAGGACAUUUGGACAAAUU (((..((((................((....)).((((.....).......(((((-(......))))))..))))))).(((((.(((..(((.....)))..)))))))))))..... ( -32.70) >DroYak_CAF1 156047 120 - 1 GUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGUCGUCCCAAUAGCGGCGAAGGCGAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUU (((((.(((...............(((....)))..((.....))...)))....((((...((((((((.((((((.......)))))).))).)))))...))))...)))))..... ( -35.30) >consensus GUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAACAAAUCGUUUGUGCUCGCUGCAAGGACAUUUGGACAAAUU (((.....................(((....)))((((.....).......((((((.......))))))..))).....(((((.(((..(((.....)))..)))))))))))..... (-29.28 = -29.44 + 0.16)


| Location | 17,265,055 – 17,265,174 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.90 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -27.58 |
| Energy contribution | -27.74 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265055 119 - 23771897 GGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAA ((((((((((......(((........))).....-.)))))))....................(((....)))))).......(..(...((((((.......))))))..)..).... ( -32.80) >DroSec_CAF1 148569 120 - 1 GGCAGACGUUGAAAAAUCACUUUAGCAUGAACCCAAAAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCUCCCCAAUAGCGGCAAAGGCCAAAA ((((((((((......(((........))).......)))))))....................(((....))))))...............((((..((......))...))))..... ( -27.62) >DroSim_CAF1 143603 119 - 1 GGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAA ((((((((((......(((........))).....-.)))))))....................(((....)))))).......(..(...((((((.......))))))..)..).... ( -32.80) >DroEre_CAF1 151622 118 - 1 GGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAAGCCGGAAGGGGCCGAAAACCGAACGAAUGCCG-CCCAAUAGCGGCAAAGGCGAAAA ((((((((((......(((........))).....-.))))))).....................((....)).)))..........((..(((((-(......))))))....)).... ( -30.20) >DroYak_CAF1 156087 119 - 1 GGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGUCGUCCCAAUAGCGGCGAAGGCGAAAA .(((((((((......(((........))).....-.))))))).((((...............(((....)))((((........(((.....)))........))))))))))..... ( -30.09) >consensus GGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA_AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAAAACCGAACGAAUGCCGCCCCAAUAGCGGCAAAGGCCAAAA ((((((((((......(((........))).......)))))))....................(((....))))))..............((((((.......)))))).......... (-27.58 = -27.74 + 0.16)

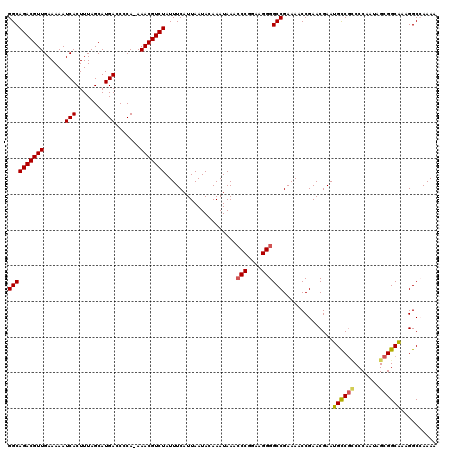
| Location | 17,265,095 – 17,265,195 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -26.65 |
| Energy contribution | -26.28 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265095 100 + 23771897 UUCGGCCCCUUCCGGGUUUAUUUGUAUUAAUGAAAUAGACGUUU-UGGGGUCAUGCUAAAGUGAUUUUUCAACGUCUGCCA------------GAAGCAGCCGGGUGGUGUGG ((((((..((((.((.((((((......)))))).((((((((.-.((((((((......))))))))..)))))))))).------------))))..))))))........ ( -32.30) >DroSec_CAF1 148609 97 + 1 UUCGGCCCCUUCCGGGUUUAUUUGUAUUAAUGAAAUAGACGUUUUUGGGUUCAUGCUAAAGUGAUUUUUCAACGUCUGCCA------------GAAGCAGCCGAGUGGU---- ((((((..((((.((.((((((......)))))).((((((((...(((.((((......)))).)))..)))))))))).------------))))..))))))....---- ( -28.60) >DroSim_CAF1 143643 96 + 1 UUCGGCCCCUUCCGGGUUUAUUUGUAUUAAUGAAAUAGACGUUU-UGGGGUCAUGCUAAAGUGAUUUUUCAACGUCUGCCA------------GAAGCAGCCGAGUGGU---- ((((((..((((.((.((((((......)))))).((((((((.-.((((((((......))))))))..)))))))))).------------))))..))))))....---- ( -33.30) >DroYak_CAF1 156127 110 + 1 UUCGGCCCCUUCCGGGUUUAUUUGUAUUAAUGAAAUAGACGUUU-UGGGGUCAUGCUAAAGUGAUUUUUCAACGUCUGCCACAAGCAGGGGCAGAAGCAGUUGGCUGGAGG-- (((.((((((.(..((((((((......)))))).((((((((.-.((((((((......))))))))..))))))))))....).)))))).))).(((....)))....-- ( -34.30) >consensus UUCGGCCCCUUCCGGGUUUAUUUGUAUUAAUGAAAUAGACGUUU_UGGGGUCAUGCUAAAGUGAUUUUUCAACGUCUGCCA____________GAAGCAGCCGAGUGGU____ .(((((..((((....((((((......)))))).((((((((...((((((((......))))))))..))))))))...............))))..)))))......... (-26.65 = -26.28 + -0.37)

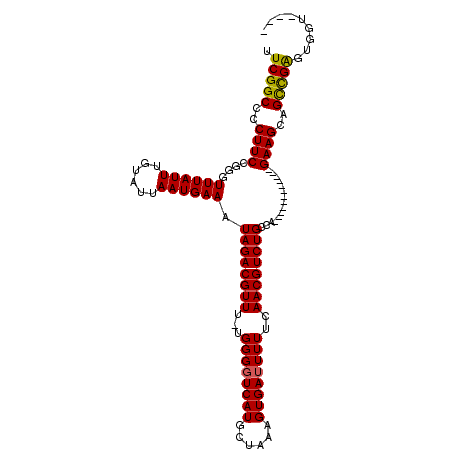
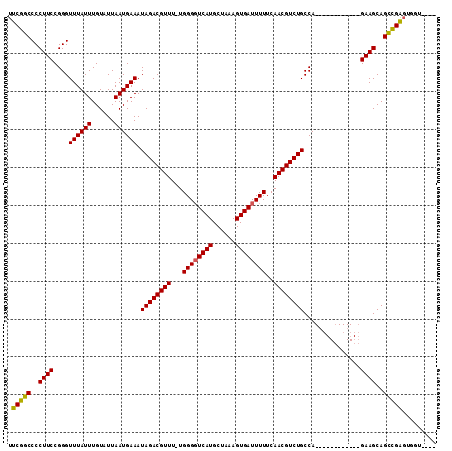
| Location | 17,265,095 – 17,265,195 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 88.43 |
| Mean single sequence MFE | -28.05 |
| Consensus MFE | -21.67 |
| Energy contribution | -21.67 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17265095 100 - 23771897 CCACACCACCCGGCUGCUUC------------UGGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAA ..........(((((.((((------------(((.(((((((......(((........))).....-.)))))))(((.....)))..........))))))).))))).. ( -28.20) >DroSec_CAF1 148609 97 - 1 ----ACCACUCGGCUGCUUC------------UGGCAGACGUUGAAAAAUCACUUUAGCAUGAACCCAAAAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAA ----.....((((((.((((------------(((.(((((((......(((........))).......)))))))(((.....)))..........))))))).)))))). ( -29.02) >DroSim_CAF1 143643 96 - 1 ----ACCACUCGGCUGCUUC------------UGGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAA ----.....((((((.((((------------(((.(((((((......(((........))).....-.)))))))(((.....)))..........))))))).)))))). ( -29.60) >DroYak_CAF1 156127 110 - 1 --CCUCCAGCCAACUGCUUCUGCCCCUGCUUGUGGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA-AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAA --......((((...((..........))...))))(((((((......(((........))).....-.)))))))....................(((....)))...... ( -25.40) >consensus ____ACCACCCGGCUGCUUC____________UGGCAGACGUUGAAAAAUCACUUUAGCAUGACCCCA_AAACGUCUAUUUCAUUAAUACAAAUAAACCCGGAAGGGGCCGAA ................................(((((((((((......(((........))).......)))))))....................(((....))))))).. (-21.67 = -21.67 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:22:02 2006