
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
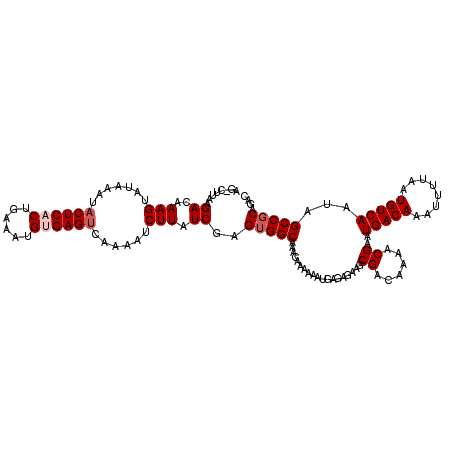
| Location | 17,264,776 – 17,264,895 |
| Length | 119 |
| Max. P | 0.894888 |

| Location | 17,264,776 – 17,264,895 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -17.54 |
| Energy contribution | -18.34 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17264776 119 + 23771897 AG-CUCAAGACAAAGUAUAAAUACUCACUGAAAUGUGAGUCAAAAUCUUAUCGAGUGGCAAACAAAAAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC ..-.....((..(((.......((((((......))))))......))).))..(((((...................((......))..(((((........)))))...))))).... ( -21.32) >DroSec_CAF1 148306 119 + 1 AG-CUUAAGACAAAGUAUAAAUACUCACUGAAAUGUGAGUCAAAAUCUUAUCGAGUGGCAAACAAAAAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC .(-(((......))))......((((((......))))))..............(((((...................((......))..(((((........)))))...))))).... ( -21.80) >DroSim_CAF1 143340 119 + 1 AG-CUUAAGACAAAGUAUAAAUACUCACUGAAAUGUGAGUCAAAAUCUUAUCGAGUGGCAAACAAAAAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC .(-(((......))))......((((((......))))))..............(((((...................((......))..(((((........)))))...))))).... ( -21.80) >DroEre_CAF1 151361 115 + 1 AAUAGUAAGACAAAGUAUAAAUACUC-CUGAAAUGCGAGCCAAAAUCUUAUCGAGAGGCAAA----AAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC ..........((.((((....)))).-.))...((((.((.....(((((((....).....----..))))))................(((((........)))))...))))))... ( -14.71) >DroYak_CAF1 155826 115 + 1 AG-CCUAAGACAAAGUAUAAAUACUC-CUGAAAUGUGAGUCGGAAUCUUAUCGAGUGGCAAA---AAAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC ..-..(((((...((((....)))).-((((........))))..)))))....(((((...---.............((......))..(((((........)))))...))))).... ( -18.10) >consensus AG_CUUAAGACAAAGUAUAAAUACUCACUGAAAUGUGAGUCAAAAUCUUAUCGAGUGGCAAACAAAAAAUGAGAGAAGCGACAAAACGAAUGACAAAUUUUAAUGUCAAUAGCCGCAGAC ........((..(((.......((((((......))))))......))).))..(((((...................((......))..(((((........)))))...))))).... (-17.54 = -18.34 + 0.80)


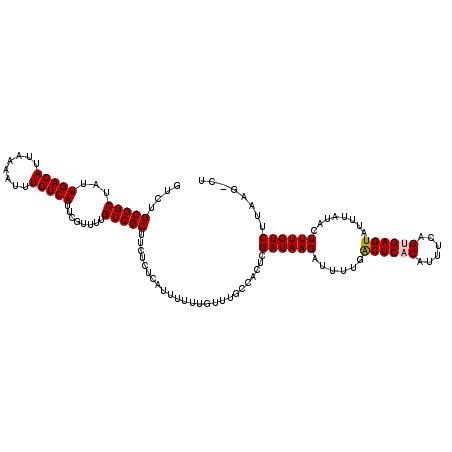
| Location | 17,264,776 – 17,264,895 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -21.91 |
| Consensus MFE | -19.74 |
| Energy contribution | -19.98 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17264776 119 - 23771897 GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUUUUUGUUUGCCACUCGAUAAGAUUUUGACUCACAUUUCAGUGAGUAUUUAUACUUUGUCUUGAG-CU ......((((...((((.((((((((.....(((....((.((......((.....))...)).)).)))..))))))))((((((......))))))..........))))...))-)) ( -23.00) >DroSec_CAF1 148306 119 - 1 GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUUUUUGUUUGCCACUCGAUAAGAUUUUGACUCACAUUUCAGUGAGUAUUUAUACUUUGUCUUAAG-CU ......((((...((((.((((((((.....(((....((.((......((.....))...)).)).)))..))))))))((((((......))))))..........))))...))-)) ( -21.90) >DroSim_CAF1 143340 119 - 1 GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUUUUUGUUUGCCACUCGAUAAGAUUUUGACUCACAUUUCAGUGAGUAUUUAUACUUUGUCUUAAG-CU ......((((...((((.((((((((.....(((....((.((......((.....))...)).)).)))..))))))))((((((......))))))..........))))...))-)) ( -21.90) >DroEre_CAF1 151361 115 - 1 GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUU----UUUGCCUCUCGAUAAGAUUUUGGCUCGCAUUUCAG-GAGUAUUUAUACUUUGUCUUACUAUU ...((((((((.(((((........)))))...(((((((((.......((...----..)).....)))))))))..)))).)))).....(-(((((....))))))........... ( -22.20) >DroYak_CAF1 155826 115 - 1 GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUUU---UUUGCCACUCGAUAAGAUUCCGACUCACAUUUCAG-GAGUAUUUAUACUUUGUCUUAGG-CU ....(((((...(((((........)))))........)))))............---...(((....((((((......((((.(......)-))))........))))))...))-). ( -20.54) >consensus GUCUGCGGCUAUUGACAUUAAAAUUUGUCAUUCGUUUUGUCGCUUCUCUCAUUUUUUGUUUGCCACUCGAUAAGAUUUUGACUCACAUUUCAGUGAGUAUUUAUACUUUGUCUUAAG_CU ....(((((...(((((........)))))........))))).........................((((((......((((((......))))))........))))))........ (-19.74 = -19.98 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:57 2006