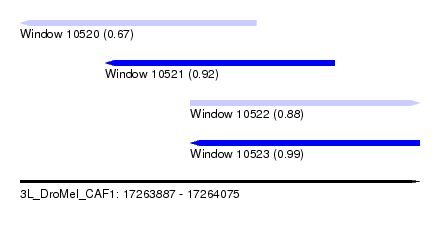
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,263,887 – 17,264,075 |
| Length | 188 |
| Max. P | 0.994906 |

| Location | 17,263,887 – 17,263,998 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 98.65 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -33.83 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.74 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263887 111 - 23771897 GGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACAACGGCAAACACCCGAAAGACUCAAAAGCGAGCGCGGCUAA ((((....))))(((((..(((((((......(((.......((((..(............)..))))....(((.......)))...))).....)).))))).))))). ( -32.80) >DroSec_CAF1 147431 111 - 1 GGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACAACGGCAAACACCCGAAAGACUCAAAAGCGAGCGCGGCUAA ((((....))))(((((..(((((((......(((.......((((..(............)..))))....(((.......)))...))).....)).))))).))))). ( -32.80) >DroSim_CAF1 142455 111 - 1 GGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACAACGGCAAACACCCGAAAGACUCAAAAGCGAGCGCGGCUAA ((((....))))(((((..(((((((......(((.......((((..(............)..))))....(((.......)))...))).....)).))))).))))). ( -32.80) >DroEre_CAF1 150586 111 - 1 GGGCGAUAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACAACGGCAAACACCCGAAAGACUCAAAAGCGAGCGCGGCCAA ((((....))))(((((..(((((((......(((.......((((..(............)..))))....(((.......)))...))).....)).))))).))))). ( -35.40) >consensus GGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACAACGGCAAACACCCGAAAGACUCAAAAGCGAGCGCGGCUAA ((((....))))(((((..(((((((......(((.......((((..(............)..))))....(((.......)))...))).....)).))))).))))). (-33.83 = -33.45 + -0.38)


| Location | 17,263,927 – 17,264,035 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -36.00 |
| Energy contribution | -35.62 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263927 108 - 23771897 AGCGAAAAGACUGGCCUUUGGAUGGGGCUGGAGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACA (((((.....(..(((((.....)))))..).(((..((((....))))..))).)))))..((((...(((((....))))).......(((.....)))))))... ( -39.60) >DroSec_CAF1 147471 107 - 1 AGCGAAAAGACUGGCCUU-UGGUGGGGCUCGGGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACA (((((.....((((((((-....))))).)))(((..((((....))))..))).)))))..((((...(((((....))))).......(((.....)))))))... ( -38.40) >DroSim_CAF1 142495 108 - 1 AGCGAAAAGACUGGCCUUUUGAUGGGGCUCGGGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACA (((((.....((((((((.....))))).)))(((..((((....))))..))).)))))..((((...(((((....))))).......(((.....)))))))... ( -38.40) >DroEre_CAF1 150626 107 - 1 AGCGGAAAGACUGGCCUUUGGAUGGGGUU-GAGGUUGGGGCGAUAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACA ...........(((((((((...(((((.-(((((..((((....))))..))).))))))).......(((((....)))))..))))..((.....)))))))... ( -36.70) >consensus AGCGAAAAGACUGGCCUUUGGAUGGGGCUCGAGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAAUGUCAUCAAAGUGGCUACGAAAUGCAUAAAGCGGCCAACA ...........(((((.......(((((....(((..((((....))))..)))...)))))..(((..(((((....)))))))).....((.....)))))))... (-36.00 = -35.62 + -0.38)


| Location | 17,263,967 – 17,264,075 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -29.91 |
| Consensus MFE | -27.63 |
| Energy contribution | -27.88 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263967 108 + 23771897 UUACCAGGAGCGAUGGCCAGAGCUUUCACCCCAACCUCCAGCCCCAUCCAAAGGCCAGUCUUUUCGCUCCGUCAUGGCUUAAUGGCAUUACGCCCAAGGCCCCACAUU ......(((((((.(((..(((.((.......)).)))..)))......(((((....))))))))))))((...(((((...(((.....)))..)))))..))... ( -29.90) >DroSec_CAF1 147511 107 + 1 UUACCAGGAGCGAUGGCCAGAGCUUUCACCCCAACCCCGAGCCCCACCA-AAGGCCAGUCUUUUCGCUCCGUCAUGGCUUAAUGGCAUUACGCCCAAGGCCCCACAUU ......((((((((((((.(.((((.............)))).).....-..)))))......)))))))((...(((((...(((.....)))..)))))..))... ( -29.32) >DroSim_CAF1 142535 108 + 1 UUACCAGGAGCGAUGGCCAGAGCUUUCACCCCAACCCCGAGCCCCAUCAAAAGGCCAGUCUUUUCGCUCCGUCAUGGCUUAAUGGCAUUACGCCCAAGGCCCCACAUU ......((((((((((((.(......)...........((......))....)))))......)))))))((...(((((...(((.....)))..)))))..))... ( -30.80) >DroEre_CAF1 150666 107 + 1 UUACCAGGAGCGAUGGCCAGAGCUAUCGCCCCAACCUC-AACCCCAUCCAAAGGCCAGUCUUUCCGCUCCGUCAUGGCUUAAUGGCAUUACGCCCAAGGCCCCACAUU ......(((((((((((....))))))(((........-.............)))..........)))))((...(((((...(((.....)))..)))))..))... ( -29.60) >consensus UUACCAGGAGCGAUGGCCAGAGCUUUCACCCCAACCCCGAGCCCCAUCAAAAGGCCAGUCUUUUCGCUCCGUCAUGGCUUAAUGGCAUUACGCCCAAGGCCCCACAUU ......((((((((((((..................................)))))......)))))))((...(((((...(((.....)))..)))))..))... (-27.63 = -27.88 + 0.25)

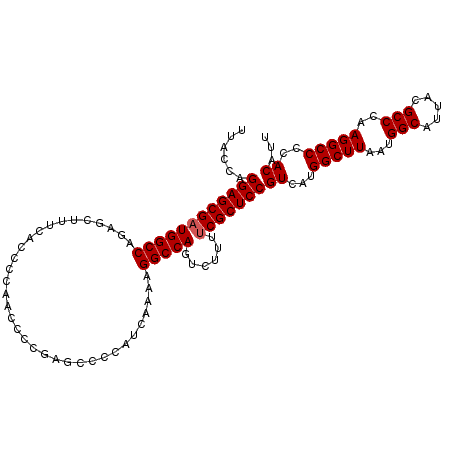
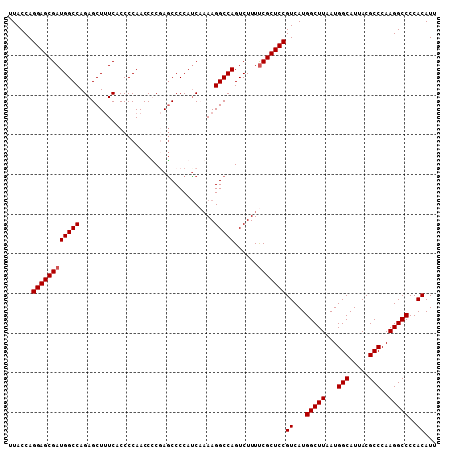
| Location | 17,263,967 – 17,264,075 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.22 |
| Mean single sequence MFE | -45.80 |
| Consensus MFE | -44.39 |
| Energy contribution | -43.82 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263967 108 - 23771897 AAUGUGGGGCCUUGGGCGUAAUGCCAUUAAGCCAUGACGGAGCGAAAAGACUGGCCUUUGGAUGGGGCUGGAGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAA ...((.((((....(((.....))).....))).).))(((((((.....(..(((((.....)))))..).(((..((((....))))..))).)))))))...... ( -48.00) >DroSec_CAF1 147511 107 - 1 AAUGUGGGGCCUUGGGCGUAAUGCCAUUAAGCCAUGACGGAGCGAAAAGACUGGCCUU-UGGUGGGGCUCGGGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAA ...((.((((....(((.....))).....))).).))(((((((.....((((((((-....))))).)))(((..((((....))))..))).)))))))...... ( -46.80) >DroSim_CAF1 142535 108 - 1 AAUGUGGGGCCUUGGGCGUAAUGCCAUUAAGCCAUGACGGAGCGAAAAGACUGGCCUUUUGAUGGGGCUCGGGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAA ...((.((((....(((.....))).....))).).))(((((((.....((((((((.....))))).)))(((..((((....))))..))).)))))))...... ( -46.80) >DroEre_CAF1 150666 107 - 1 AAUGUGGGGCCUUGGGCGUAAUGCCAUUAAGCCAUGACGGAGCGGAAAGACUGGCCUUUGGAUGGGGUU-GAGGUUGGGGCGAUAGCUCUGGCCAUCGCUCCUGGUAA ...((.((((....(((.....))).....))).).))(((((((.......((((((.....))))))-..(((..((((....))))..))).)))))))...... ( -41.60) >consensus AAUGUGGGGCCUUGGGCGUAAUGCCAUUAAGCCAUGACGGAGCGAAAAGACUGGCCUUUGGAUGGGGCUCGAGGUUGGGGUGAAAGCUCUGGCCAUCGCUCCUGGUAA ...((.((((....(((.....))).....))).).))(((((((.....(.((((((.....)))))).).(((..((((....))))..))).)))))))...... (-44.39 = -43.82 + -0.56)

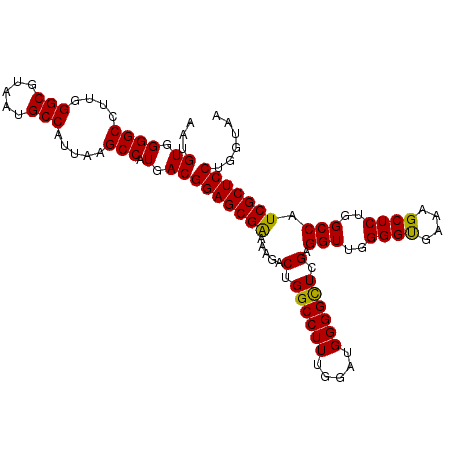

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:51 2006