
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,263,155 – 17,263,437 |
| Length | 282 |
| Max. P | 0.998962 |

| Location | 17,263,155 – 17,263,252 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 87.60 |
| Mean single sequence MFE | -17.56 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263155 97 + 23771897 ACCUCUUCGUACUCCCUCCUUCU----UUAUUUAU--UUUUUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUU ........(((............----........--..(((((((.((((((((...)).)))))).))))))).((.(((((....))))))).))).... ( -17.40) >DroSec_CAF1 146719 98 + 1 UCCUCUUCGUACUCCUUCUUCCUC--CUCUUCUAU---UUUUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUU ........(((.............--.........---.(((((((.((((((((...)).)))))).))))))).((.(((((....))))))).))).... ( -17.40) >DroSim_CAF1 141730 98 + 1 UCCUCUUCGCACUCCCUCUUCCUC--CUUUUCUAU---UUUUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUU ........(((.............--.........---.(((((((.((((((((...)).)))))).))))))).((.(((((....))))))).))).... ( -19.40) >DroYak_CAF1 154167 103 + 1 UCCUUUUCAAACUCUCUCUCUCUCUCUCAUUCUAUAUAUUUUUUGCGAUUUAAACGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUU ........................((((((.((((....................)))).))))))((((((((...((....))..))))))))........ ( -16.05) >consensus UCCUCUUCGUACUCCCUCUUCCUC__CUUUUCUAU___UUUUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUU ...........................................(((.((......)).)))((..(((((((((...((....))..)))))))))..))... (-15.60 = -15.60 + -0.00)

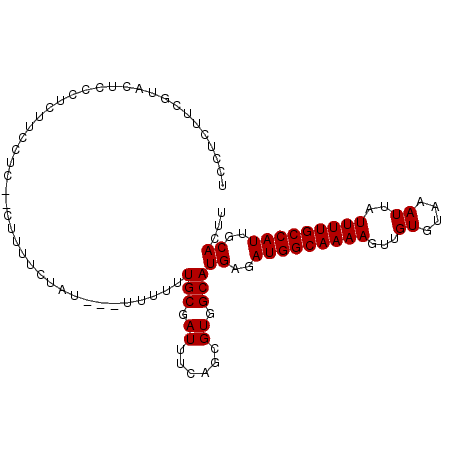

| Location | 17,263,189 – 17,263,292 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -25.58 |
| Energy contribution | -25.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263189 103 + 23771897 UUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCCUUU ....((.......)).(((((((..(((((((((...((....))..)))))))))..))..)))))((((((...((....))....))))))......... ( -28.30) >DroSec_CAF1 146754 103 + 1 UUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUU ....((.......)).(((((((..(((((((((...((....))..)))))))))..))..)))))((((((...((....))....))))))......... ( -28.30) >DroSim_CAF1 141765 103 + 1 UUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUU ....((.......)).(((((((..(((((((((...((....))..)))))))))..))..)))))((((((...((....))....))))))......... ( -28.30) >DroYak_CAF1 154207 103 + 1 UUUUGCGAUUUAAACGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCUCAACUUCCUUUUGGUCCUUUGUGCAUCUCUUUU ......(((.......(((((((..(((((((((...((....))..)))))))))..))..)))))((.(((...((....))....))).)))))...... ( -23.20) >consensus UUUUGCGAUUUCAGCGUGGCAUGAGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUU ................(((((((..(((((((((...((....))..)))))))))..))..)))))((((((...((....))....))))))......... (-25.58 = -25.82 + 0.25)

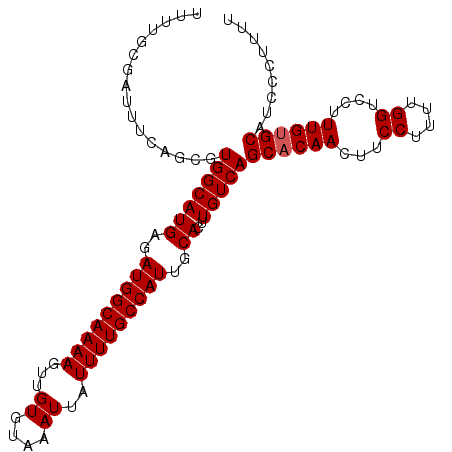

| Location | 17,263,189 – 17,263,292 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 97.57 |
| Mean single sequence MFE | -23.09 |
| Consensus MFE | -21.55 |
| Energy contribution | -21.79 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263189 103 - 23771897 AAAGGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCUCAUGCCACGCUGAAAUCGCAAAA ....(.(((((((((....((....))...))))))...((.((((((((((((((.............)))))))))....)).)))..))..))).).... ( -23.52) >DroSec_CAF1 146754 103 - 1 AAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCUCAUGCCACGCUGAAAUCGCAAAA ....(.(((((((((....((....))...))))))...((.((((((((((((((.............)))))))))....)).)))..))..))).).... ( -23.72) >DroSim_CAF1 141765 103 - 1 AAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCUCAUGCCACGCUGAAAUCGCAAAA ....(.(((((((((....((....))...))))))...((.((((((((((((((.............)))))))))....)).)))..))..))).).... ( -23.72) >DroYak_CAF1 154207 103 - 1 AAAAGAGAUGCACAAAGGACCAAAAGGAAGUUGAGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCUCAUGCCACGUUUAAAUCGCAAAA ....(((((((((......((....))..(((.....)))..))))).((((((((.............)))))))))))).(((............)))... ( -21.42) >consensus AAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCUCAUGCCACGCUGAAAUCGCAAAA ....(.(((((((((....((....))...))))))......((((((((((((((.............)))))))))....)).)))......))).).... (-21.55 = -21.79 + 0.25)

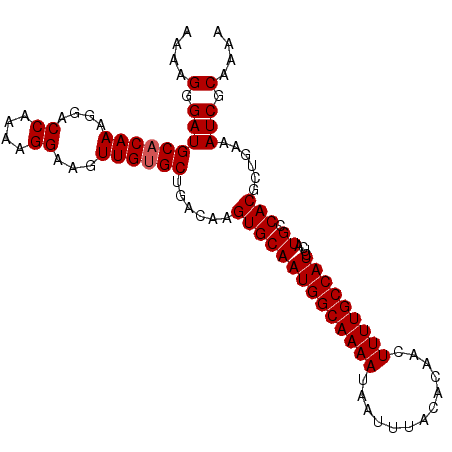

| Location | 17,263,212 – 17,263,332 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.38 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263212 120 + 23771897 AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCCUUUGACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAU .((((((((((...((....))..))))))))))(((((.((..((((((...((....))....)))))).......(((((..........)))))..)).)))))............ ( -32.10) >DroSec_CAF1 146777 120 + 1 AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUUGACACACUUUGACGUCAGUUGCCAGUGCACACCAGCUAAU .((((((((((...((....))..))))))))))(((((.((..((((((...((....))....)))))).......(((((..........)))))..)).)))))............ ( -32.10) >DroSim_CAF1 141788 120 + 1 AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUUGACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAU .((((((((((...((....))..))))))))))(((((.((..((((((...((....))....)))))).......(((((..........)))))..)).)))))............ ( -32.10) >DroEre_CAF1 149889 120 + 1 AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCCCAACUUCCUUUUGGUCCUUUGUGCGUCUCUUUUGACACACUUUGACGUCAGUUGCCAGUGCAGAGCGGCUAAU .((((((((((...((....))..))))))))))(((((.(.((((.(((.......)))(((....(((.(((......))).)))...)))....))))).)))))............ ( -32.50) >DroYak_CAF1 154230 120 + 1 AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCUCAACUUCCUUUUGGUCCUUUGUGCAUCUCUUUUGACACACUUUGACGUCAGUUGCCAGUGCGGAGCGGCUAAU .((((((((((...((....))..))))))))))((........))...........(((((((((((..(....(..(((((..........)))))..)...)..))))).)))))). ( -29.80) >consensus AGAUGGCAAAAGUUGUGUAAAUUAUUUUGCCAUUGCACUUGUCAGCACAACUUCCUUUUGGUCCUUUGUGCAUCCCUUUUGACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAU .((((((((((...((....))..))))))))))(((((.((..((((((...((....))....)))))).......(((((..........)))))..)).)))))............ (-29.98 = -30.38 + 0.40)

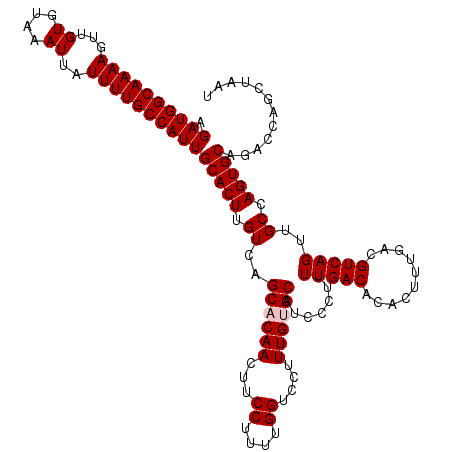

| Location | 17,263,212 – 17,263,332 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -30.28 |
| Energy contribution | -30.12 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263212 120 - 23771897 AUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAGGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ......((((((....((((.......))))..(((((((......)))))))...))))))...(((.((((..((....))..))))(((((((.............))))))).))) ( -33.42) >DroSec_CAF1 146777 120 - 1 AUUAGCUGGUGUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ....((..((....))..))..((...(((...(((((((......)))))))....)))....))...((((..((....))..))))(((((((.............))))))).... ( -32.62) >DroSim_CAF1 141788 120 - 1 AUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ......((((((....((((.......))))..(((((((......)))))))...))))))...(((.((((..((....))..))))(((((((.............))))))).))) ( -33.42) >DroEre_CAF1 149889 120 - 1 AUUAGCCGCUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGAGACGCACAAAGGACCAAAAGGAAGUUGGGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ...........((((((..((.(..((......(((((((......)))))))......((....))..))..)..))...))))))(((((((((.............))))))))).. ( -34.92) >DroYak_CAF1 154230 120 - 1 AUUAGCCGCUCCGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGAGAUGCACAAAGGACCAAAAGGAAGUUGAGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ............((((((....)....((((..(((((((......)))))))......((....)).........)))).))))).(((((((((.............))))))))).. ( -29.92) >consensus AUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGACAAGUGCAAUGGCAAAAUAAUUUACACAACUUUUGCCAUCU ............((((((....)....((((..(((((((......)))))))......((....)).........)))).))))).(((((((((.............))))))))).. (-30.28 = -30.12 + -0.16)



| Location | 17,263,252 – 17,263,372 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -36.04 |
| Energy contribution | -36.88 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263252 120 - 23771897 UUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAGGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGAC ....(..(((((..(..(((....)))..).(((((((......))((((((....((((.......))))..(((((((......)))))))...)))))).))))))))))..).... ( -43.00) >DroSec_CAF1 146817 120 - 1 UUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUGUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGAC ....(..(((((..(..(((....)))..).(((((........((..((....))..)).......(((...(((((((......)))))))....)))...))))))))))..).... ( -41.30) >DroSim_CAF1 141828 120 - 1 UUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGAC ....(..(((((..(..(((....)))..).(((((((......))((((((....((((.......))))..(((((((......)))))))...)))))).))))))))))..).... ( -43.00) >DroEre_CAF1 149929 120 - 1 UUUUGUGAAGCUGUUCGCCGGAAACGCUGGUUCCUUGCAAAUUAGCCGCUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGAGACGCACAAAGGACCAAAAGGAAGUUGGGCUGAC .(((((.(((....(((.((....)).)))...))))))))((((((((...)).....(((((..((((...(((((((......)))))))....))).....)..))))))))))). ( -34.00) >DroYak_CAF1 154270 120 - 1 UUUUGCGAAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCCGCUCCGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGAGAUGCACAAAGGACCAAAAGGAAGUUGAGCUGAC .(((((((((((....((((....))))))))..)))))))(((((((((((((....)).......(((...(((((((......)))))))....))).....)).)).)).))))). ( -37.90) >consensus UUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUCAAAAGGGAUGCACAAAGGACCAAAAGGAAGUUGUGCUGAC ....((((((((....((((....))))((.(((((........((((((....)))))).............(((((((......))))))).))))))).......)))))))).... (-36.04 = -36.88 + 0.84)


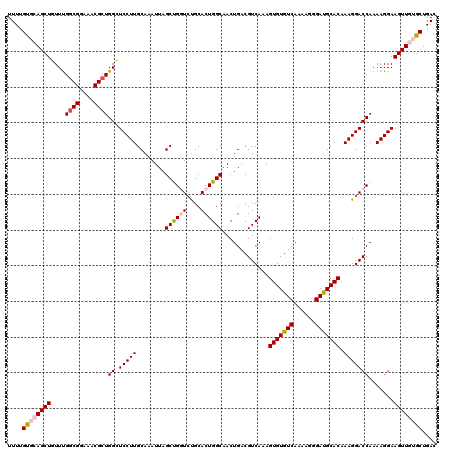
| Location | 17,263,292 – 17,263,412 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -33.10 |
| Energy contribution | -32.62 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263292 120 + 23771897 GACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAUUUGCAAGGAGCCAGCGUUUCCGCCAAACAGCUGCACAAAAUUGAUGAAUAACAAGCUGGGGCACUGGAAAAACAGUAUUA (((..........))).(((.(((((((...((((((.((((((((.(.((.(((((((.....)))).))))).)....))).)))))....)))))).)))))))...)))....... ( -34.90) >DroSec_CAF1 146857 120 + 1 GACACACUUUGACGUCAGUUGCCAGUGCACACCAGCUAAUUUGCAAGGAGCCAGCGUUUCCGCCAAACAGCUGCACAAAAUUGAUGAAUAACAAGCUGGGGCACUGGAAAAACAGUAUUA (((..........))).(((.(((((((...((((((.((((((((.(.((.(((((((.....)))).))))).)....))).)))))....)))))).)))))))...)))....... ( -34.90) >DroSim_CAF1 141868 120 + 1 GACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAUUUGCAAGGAGCCAGCGUUUCCGCCAAACAGCUGCACAAAAUUGAUGAAUAACAAGCUGGGGCACUGGAAAAACAGUAUUA (((..........))).(((.(((((((...((((((.((((((((.(.((.(((((((.....)))).))))).)....))).)))))....)))))).)))))))...)))....... ( -34.90) >DroEre_CAF1 149969 120 + 1 GACACACUUUGACGUCAGUUGCCAGUGCAGAGCGGCUAAUUUGCAAGGAACCAGCGUUUCCGGCGAACAGCUUCACAAAAUUGAUGAAUAACAAGCUGCGGCACUGGAAAAACAGUAUUA (((..........))).(((.(((((((...((((((..(((((..(((((....)))))..)))))....((((((....)).)))).....)))))).)))))))...)))....... ( -37.20) >DroYak_CAF1 154310 120 + 1 GACACACUUUGACGUCAGUUGCCAGUGCGGAGCGGCUAAUUUGCAAGGAGCCAGCGUUUCCGCCAAACAGCUUCGCAAAAUUGAUGAAUAACAAGCUGCGGCACUGGAAAAACAGUAUUA (((..........))).(((.((((((((((((((((..(.....)..)))).(((....)))......)))))(((...(((........)))..))).)))))))...)))....... ( -35.60) >consensus GACACACUUUGACGUCAGUUGCCAGUGCAGACCAGCUAAUUUGCAAGGAGCCAGCGUUUCCGCCAAACAGCUGCACAAAAUUGAUGAAUAACAAGCUGGGGCACUGGAAAAACAGUAUUA (((..........))).(((.(((((((...((((((.((((((((.....((((((((.....)))).)))).......))).)))))....)))))).)))))))...)))....... (-33.10 = -32.62 + -0.48)


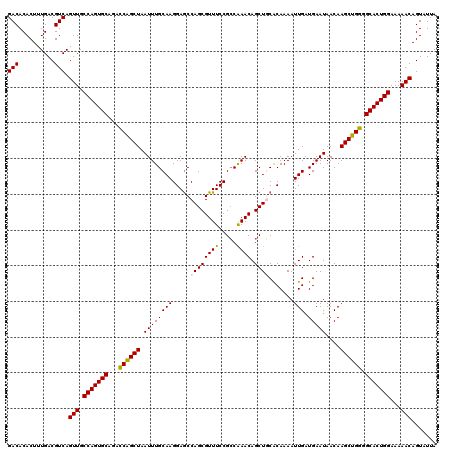
| Location | 17,263,292 – 17,263,412 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.60 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263292 120 - 23771897 UAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...(((((((.((((((.(((...((.......))(((((..(.(..(((....)))..))...)))))))).))))))...))))))).)))((....)).)))))... ( -37.20) >DroSec_CAF1 146857 120 - 1 UAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUGUGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...((((((((((((((.(((...((.......))(((((..(.(..(((....)))..))...)))))))).)))))).).))))))).)))((....)).)))))... ( -38.90) >DroSim_CAF1 141868 120 - 1 UAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...(((((((.((((((.(((...((.......))(((((..(.(..(((....)))..))...)))))))).))))))...))))))).)))((....)).)))))... ( -37.20) >DroEre_CAF1 149969 120 - 1 UAAUACUGUUUUUCCAGUGCCGCAGCUUGUUAUUCAUCAAUUUUGUGAAGCUGUUCGCCGGAAACGCUGGUUCCUUGCAAAUUAGCCGCUCUGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...(((((((.((.(((.(((.(((((.......)))))((.....(((((.....))))).....)).))).))).))...))))))).)))((....)).)))))... ( -29.80) >DroYak_CAF1 154310 120 - 1 UAAUACUGUUUUUCCAGUGCCGCAGCUUGUUAUUCAUCAAUUUUGCGAAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCCGCUCCGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...(((((((.(.(((..((((.........(((((((((((....((((....))))))))..))))))).)))).)))).))))))).)))((....)).)))))... ( -35.50) >consensus UAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAAAUUAGCUGGUCUGCACUGGCAACUGACGUCAAAGUGUGUC ..((((((((...(((((((.((((((((........))).((((((.((((....((((....))))))))...))))))...)))))...))))))).)))((....)).)))))... (-32.80 = -32.60 + -0.20)

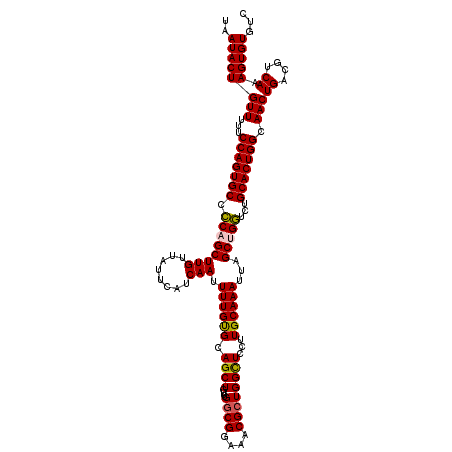
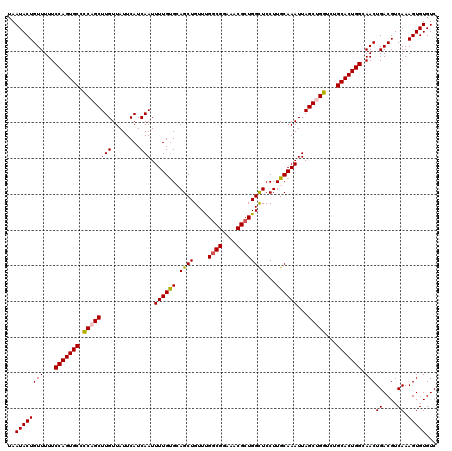
| Location | 17,263,332 – 17,263,437 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -20.70 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17263332 105 - 23771897 UCUUA---------------AGAUUGUUAUUCCAAUGCACUAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAA ...((---------------((((((..((..(((.((.....(((((......))))).....)))))..))....))))))))(((((..(.(..(((....)))..))...))))). ( -25.70) >DroSec_CAF1 146897 98 - 1 ----------------------GUUGUUAUUCCAAUGCACUAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAA ----------------------.............((((.....((((......))))(((.(((((.((....((.......)).)))))))...((((....)))))))....)))). ( -24.50) >DroSim_CAF1 141908 105 - 1 UCUUA---------------AGAUUGUUAUUCCAAUGCACUAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAA ...((---------------((((((..((..(((.((.....(((((......))))).....)))))..))....))))))))(((((..(.(..(((....)))..))...))))). ( -25.70) >DroEre_CAF1 150009 120 - 1 CCUUGGGAAACUUUCCCGCAAAACUGUUAUUUCAGUGCACUAAUACUGUUUUUCCAGUGCCGCAGCUUGUUAUUCAUCAAUUUUGUGAAGCUGUUCGCCGGAAACGCUGGUUCCUUGCAA ....((((.....))))((((.((((..(...(((((......)))))...)..))))(((((((((......((((.......))))))))))..((.(....))).)))...)))).. ( -30.00) >DroYak_CAF1 154350 120 - 1 UCUUGAGAAACUUUGCCGCAAAACUGUUGUUUCAAUGCACUAAUACUGUUUUUCCAGUGCCGCAGCUUGUUAUUCAUCAAUUUUGCGAAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAA ......(((((...((.(.....).)).)))))..((((.....((((......))))((((((((((((..............)).)))))))..((((....)))))))....)))). ( -30.94) >consensus UCUUA_______________AAAUUGUUAUUCCAAUGCACUAAUACUGUUUUUCCAGUGCCCCAGCUUGUUAUUCAUCAAUUUUGUGCAGCUGUUUGGCGGAAACGCUGGCUCCUUGCAA ...................................((((.....((((......))))(((.(((((.((....((.......)).)))))))...((((....)))))))....)))). (-20.70 = -21.14 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:46 2006