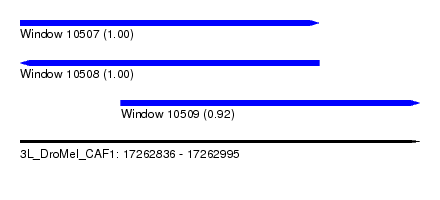
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,262,836 – 17,262,995 |
| Length | 159 |
| Max. P | 0.999196 |

| Location | 17,262,836 – 17,262,955 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -45.34 |
| Consensus MFE | -42.10 |
| Energy contribution | -42.34 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17262836 119 + 23771897 UCAUCCAGUUGGGUCGACUUGGCGGACUUGGCAUUUCAGCUCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUG .(((.((((..((((((...(.(((((..((((..((((((.........)))))).))))))))).)((((((....))))))................-.))))..))..)))).))) ( -46.10) >DroSec_CAF1 146400 119 + 1 UCAUCCAGUUGGGUCGACUUGGCGGACUUGGCAUUUCAGCUCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUG .(((.((((..((((((...(.(((((..((((..((((((.........)))))).))))))))).)((((((....))))))................-.))))..))..)))).))) ( -46.10) >DroSim_CAF1 141411 119 + 1 UCAUCCAGUUGGGUCGACUUGGCGGACUUGGCAUUUCAGCUCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUG .(((.((((..((((((...(.(((((..((((..((((((.........)))))).))))))))).)((((((....))))))................-.))))..))..)))).))) ( -46.10) >DroEre_CAF1 149493 119 + 1 UCAUCCAGUUGGGCCGACUUGGCGGACUUGGCAUUUCAGCUCUCCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCAUUUCGUGCCACUCCU-UUCGAGUCCUGGCUGCGUG .(((.((((..((...((((((.(((..((((((.((((((.........))))))..(((....)))((((((....)))))).....)))))).))).-.))))))))..)))).))) ( -46.60) >DroYak_CAF1 153850 120 + 1 UCAUCCAGUUGGCUCGACUUGGCGGACUUGGCAUUCCAGCUCUCCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGAAUCCUCCAUUGCGUACCACUCGUAGUCAAGUCCUGGCUGCGUG .(((.((((..(...((((((((((((..((((...(((((.........)))))..))))))))...(((((......)))))..((((.......)))))))))))))..)))).))) ( -41.80) >consensus UCAUCCAGUUGGGUCGACUUGGCGGACUUGGCAUUUCAGCUCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU_UUCGAGUCCUGGCUGCGUG .(((.((((..(...((((((((((((..((((..((((((.........)))))).)))))))))..((((((....))))))..................))))))))..)))).))) (-42.10 = -42.34 + 0.24)

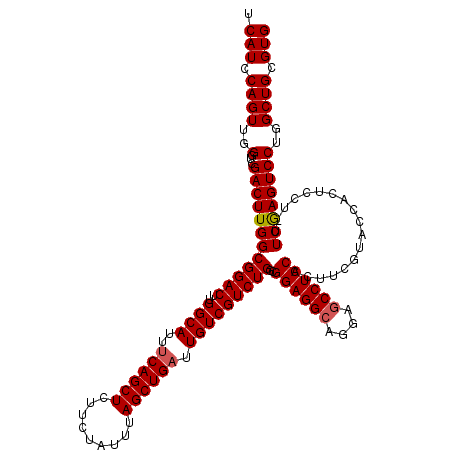
| Location | 17,262,836 – 17,262,955 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.31 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -38.36 |
| Energy contribution | -38.52 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17262836 119 - 23771897 CACGCAGCCAGGACUCGAA-AGGAGUGGUACGAAGUGGAGGCUCCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGAAGAGCUGAAAUGCCAAGUCCGCCAAGUCGACCCAACUGGAUGA ....((.((((...((((.-.(((.(((((....((((((((....))))))))..........((((((.........))))))..)))))..)))......)))).....)))).)). ( -42.00) >DroSec_CAF1 146400 119 - 1 CACGCAGCCAGGACUCGAA-AGGAGUGGUACGAAGUGGAGGCUCCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGAAGAGCUGAAAUGCCAAGUCCGCCAAGUCGACCCAACUGGAUGA ....((.((((...((((.-.(((.(((((....((((((((....))))))))..........((((((.........))))))..)))))..)))......)))).....)))).)). ( -42.00) >DroSim_CAF1 141411 119 - 1 CACGCAGCCAGGACUCGAA-AGGAGUGGUACGAAGUGGAGGCUCCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGAAGAGCUGAAAUGCCAAGUCCGCCAAGUCGACCCAACUGGAUGA ....((.((((...((((.-.(((.(((((....((((((((....))))))))..........((((((.........))))))..)))))..)))......)))).....)))).)). ( -42.00) >DroEre_CAF1 149493 119 - 1 CACGCAGCCAGGACUCGAA-AGGAGUGGCACGAAAUGGAGGCUCCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGGAGAGCUGAAAUGCCAAGUCCGCCAAGUCGGCCCAACUGGAUGA ....((.((((........-.(((.(((((......((((((....))))))............((((((.........))))))..)))))..)))(((.....)))....)))).)). ( -42.10) >DroYak_CAF1 153850 120 - 1 CACGCAGCCAGGACUUGACUACGAGUGGUACGCAAUGGAGGAUUCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGGAGAGCUGGAAUGCCAAGUCCGCCAAGUCGAGCCAACUGGAUGA ....((.((((..(((((((..(.((((.(((((..(((((......)))))............((((((.........))))))..)))...))))))).)))))))....)))).)). ( -36.90) >consensus CACGCAGCCAGGACUCGAA_AGGAGUGGUACGAAGUGGAGGCUCCUGCCUCCGCCAGACGACAAUCAGCUAAAUAGAAGAGCUGAAAUGCCAAGUCCGCCAAGUCGACCCAACUGGAUGA ....((.((((...((((...(((.(((((((..((((((((....))))))))....))....((((((.........))))))..)))))..)))......)))).....)))).)). (-38.36 = -38.52 + 0.16)

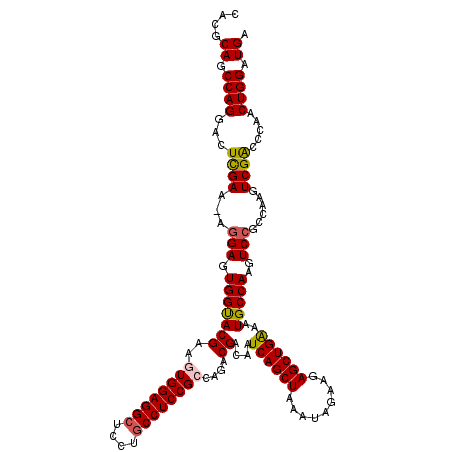
| Location | 17,262,876 – 17,262,995 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17262876 119 + 23771897 UCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUGUGCUUAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((..((.((.(((.((.((((((....))))))..........((((..-...)))))).))).))(((((.((((((....)))))))))))..))..))........ ( -32.60) >DroSec_CAF1 146440 119 + 1 UCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUGUGCUCAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((..((.((.(((.((.((((((....))))))..........((((..-...)))))).))).))(((((.((((((....)))))))))))..))..))........ ( -34.80) >DroSim_CAF1 141451 119 + 1 UCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU-UUCGAGUCCUGGCUGCGUGUGCUCAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((..((.((.(((.((.((((((....))))))..........((((..-...)))))).))).))(((((.((((((....)))))))))))..))..))........ ( -34.80) >DroEre_CAF1 149533 119 + 1 UCUCCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCAUUUCGUGCCACUCCU-UUCGAGUCCUGGCUGCGUGUGCUCAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((..((.........((((((((....)))))).......(((((((..-...))))...))).))(((((.((((((....)))))))))))..))..))........ ( -35.40) >DroYak_CAF1 153890 120 + 1 UCUCCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGAAUCCUCCAUUGCGUACCACUCGUAGUCAAGUCCUGGCUGCGUGUGCUCAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((((((....))).)))(((((......))))).((((....(((.((((((((.....)))))))).))).((((((....))))))...........))))...... ( -35.20) >consensus UCUUCUAUUUAGCUGAUUGUCGUCUGGCGGAGGCAGGAGCCUCCACUUCGUACCACUCCU_UUCGAGUCCUGGCUGCGUGUGCUCAGCAUUUUUGUUGACAUAUUCAUUGGCAAACUCAA ...........((..((.((.(((.((.((((((....))))))..........((((......)))))).))).))(((((.((((((....)))))))))))..))..))........ (-32.68 = -32.92 + 0.24)

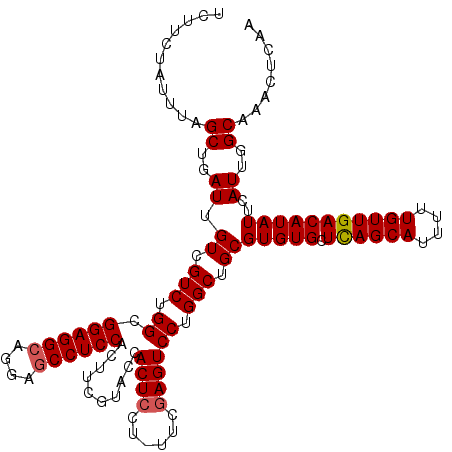
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:38 2006