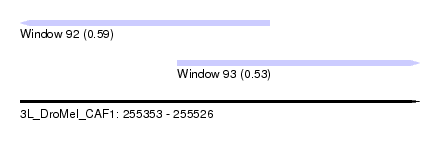
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 255,353 – 255,526 |
| Length | 173 |
| Max. P | 0.593986 |

| Location | 255,353 – 255,461 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -22.32 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 255353 108 - 23771897 GCACCCACAUUCCUUUAAAUCCGCUAGCCCAACCACUCUUAUACGGCUGUUCCACUCAACGCGAAUCCUCUCCGGACGACA--ACA--CAGGAAGGGGUGCCCUGCACACGC ((((((...(((((.......(((.((((...............))))(((......))))))..(((.....))).....--...--.))))).))))))........... ( -26.46) >DroSec_CAF1 23183 110 - 1 GCACCCACAUUCCUUUAAAUCCGCUAGCCCAACCACUCUUAUACGGCUGUUCCACUCAACGCGAAUACUCUCCGGACGACA--GCAGGAAGGAAAGGGUGCCCUGCACACGC ((((((...(((((.....((((((((((...............))))...........((.((......))))......)--)).)))))))).))))))........... ( -25.46) >DroSim_CAF1 23649 110 - 1 GCACCCACAUUCCUUUAAAUCCGCUAGCCCAACCACUCUUAUACGGCUGUUCCACUCAACGCGAAUACUCUCCGGACGACA--GCAGGAAGGAAAGGGUGCCCUGCACACGC ((((((...(((((.....((((((((((...............))))...........((.((......))))......)--)).)))))))).))))))........... ( -25.46) >DroEre_CAF1 23474 105 - 1 GCACCCACAUUCCUCCAAAUUCGCCAGCCUAACCACUCUUAUACGGCUAUUCCACUCAACGCGAAUUCUCUCCGGACGAC-------GAAGGAAGGGGUGCCCUGCACACGC ((((((...(((((...(((((((.((((...............))))............))))))).....((.....)-------).))))).))))))........... ( -26.28) >DroYak_CAF1 23342 112 - 1 GCACCCACAUUCCUCCAAAUUCGCCAGCCUAACCACUCUUAUACGGCUGUUCCACUCAACGCGAAUUCUCUCCGGACGACCAGGAAGGAAGGAAGGGGUGCCCUGCACACGC ((((((...(((((((.((((((((((((...............)))))...........)))))))...(((((....)).))).)).))))).))))))........... ( -36.66) >consensus GCACCCACAUUCCUUUAAAUCCGCUAGCCCAACCACUCUUAUACGGCUGUUCCACUCAACGCGAAUACUCUCCGGACGACA__GCAGGAAGGAAGGGGUGCCCUGCACACGC ((((((...(((((..........(((((...............)))))..........((.((......))))...............))))).))))))........... (-22.32 = -22.40 + 0.08)

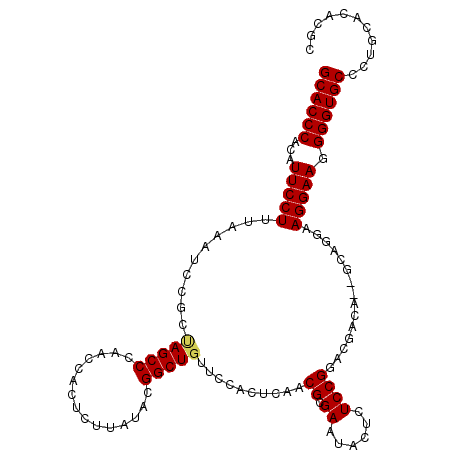
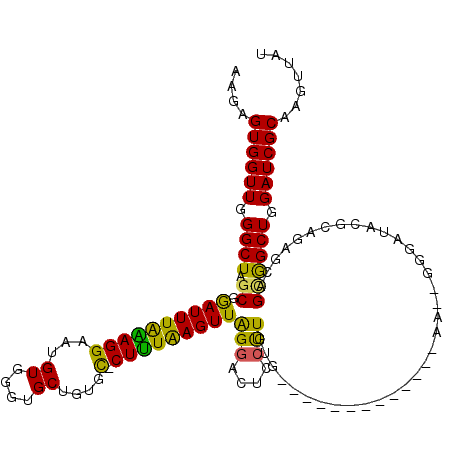
| Location | 255,421 – 255,526 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.88 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -20.48 |
| Energy contribution | -18.88 |
| Covariance contribution | -1.60 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527655 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 255421 105 + 23771897 AAGAGUGGUUGGGCUAGCGGAUUUAAAGGAAUGUGGGUGCUGUGC-CUUUAAGUUAGGACUCCUUGUG-------------ACUCGGGAUACG-AGAGAAGAGGCUGGAUCGCGAGAUAU ....((((((.(((.....((((((((((...((....))....)-)))))))))....(((.((...-------------.((((.....))-)).)).)))))).))))))....... ( -29.80) >DroSec_CAF1 23253 104 + 1 AAGAGUGGUUGGGCUAGCGGAUUUAAAGGAAUGUGGGUGCUGUGC-CUUUAAGUUAGGACUCCUUGUG-------------AA--GGGAUACGCAGAGCAGCGGCUGGAUCGCAAGUUAU ....((((((.((((..((.((((....)))).))(.((((.(((-(((......)))..(((((...-------------.)--))))...))).)))).))))).))))))....... ( -32.10) >DroSim_CAF1 23719 104 + 1 AAGAGUGGUUGGGCUAGCGGAUUUAAAGGAAUGUGGGUGCUGUGC-CUUUAAGUUAGGACUCCUUGUG-------------AA--GGGAUACGCAGAGCAGCGGCUGGAUCGCAAGUUAU ....((((((.((((..((.((((....)))).))(.((((.(((-(((......)))..(((((...-------------.)--))))...))).)))).))))).))))))....... ( -32.10) >DroEre_CAF1 23539 104 + 1 AAGAGUGGUUAGGCUGGCGAAUUUGGAGGAAUGUGGGUGCUGUGC-CUUUAAGUUGGGAUGCGCUAUG-------------GA--GGGACACGCAGACCAGUAGCUAGAUCGCAAGUUAA ....((((((.(((((.(.((((((((((...((....))....)-)))))))))((..((((...((-------------..--....))))))..)).)))))).))))))....... ( -31.60) >DroYak_CAF1 23414 118 + 1 AAGAGUGGUUAGGCUGGCGAAUUUGGAGGAAUGUGGGUGCUGUGUUCUCUGAGUUAGGAUGCCCUAUGAUAUGAGGGAUACGA--GGGAUACGCACAGCAGUAGCUAGAUCGCAAGUUAU ....((((((.(((((.((.((((....)))).))..(((((((((((((..((.......((((........))))..)).)--))))...)))))))).))))).))))))....... ( -33.40) >consensus AAGAGUGGUUGGGCUAGCGGAUUUAAAGGAAUGUGGGUGCUGUGC_CUUUAAGUUAGGACUCCUUGUG_____________AA__GGGAUACGCAGAGCAGCGGCUGGAUCGCAAGUUAU ....((((((.((((.((.((((((((((...((....))....).)))))))))(((....)))...................................)))))).))))))....... (-20.48 = -18.88 + -1.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:26 2006