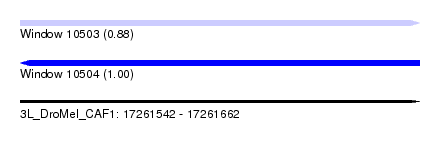
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,261,542 – 17,261,662 |
| Length | 120 |
| Max. P | 0.998181 |

| Location | 17,261,542 – 17,261,662 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -31.26 |
| Energy contribution | -33.10 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17261542 120 + 23771897 AACAGUCCCGGGCACUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAACGUCGACGAACAAGAGCCUCCAAGAAGACGAAGGAGAGCAACGCCAGUUGCGCCAGGACA ....((((..(((.((.(((((((....((((((..(((.((.......)).)))..))))))..)).))))).)).((((...........)))).(((((....)))))))).)))). ( -39.40) >DroSec_CAF1 145110 120 + 1 AACAGUCCCGGGCACUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAACGUCGACGAACAAGAGCCUCCAAGAAGACGAAGGAGAGCAACGCCAGUUGCGCCAGGACA ....((((..(((.((.(((((((....((((((..(((.((.......)).)))..))))))..)).))))).)).((((...........)))).(((((....)))))))).)))). ( -39.40) >DroSim_CAF1 140011 120 + 1 AACAGUCCCGGGCACUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAACGUCGACGAACAAGAGCCUCCAAGAAGACGAAGGAGAGCAACGCCAGUUGCGCCAGGACA ....((((..(((.((.(((((((....((((((..(((.((.......)).)))..))))))..)).))))).)).((((...........)))).(((((....)))))))).)))). ( -39.40) >DroEre_CAF1 148228 118 + 1 AAGAGUCCCGGGCACUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAACGAGGACGAACAGGAGCCUCCAAGAAGACGAAGGAGAUG--CUGCCGUUGCGCCAGUACA ....((.(.((((.(((((((((((((...))))).(((.((.......)).))))))))).))((((..(((.(..((((...........)))).).--))).)))))).)).).)). ( -28.50) >DroYak_CAF1 152510 118 + 1 AACAGUCCCGGGCGCUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAAGGACGACGAACAAGAGCCUCCAAGAAGACGAAGGAGCGC--CUGCCGUUGCGCCAGGACA ....((((..((((((((((((((.....(((((.(((.((.....))..))).)))))...))))).))))((.((((((...........)))).))--))......))))).)))). ( -38.10) >consensus AACAGUCCCGGGCACUGUUGUUGUAAUUAUGUUGCCGGCUGUAAAACAAACUGCCAACAACGUCGACGAACAAGAGCCUCCAAGAAGACGAAGGAGAGCAACGCCAGUUGCGCCAGGACA ....((((..(((.((.(((((((....((((((..(((.((.......)).)))..))))))..)).))))).)).((((...........)))).(((((....)))))))).)))). (-31.26 = -33.10 + 1.84)

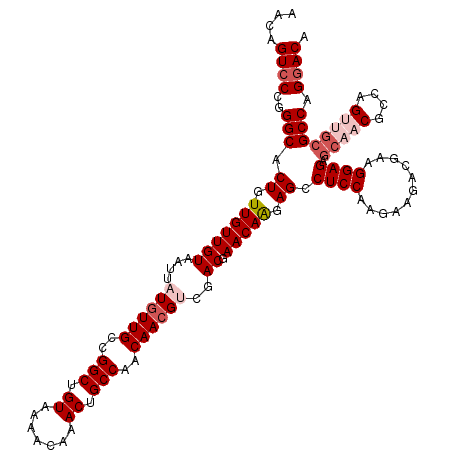
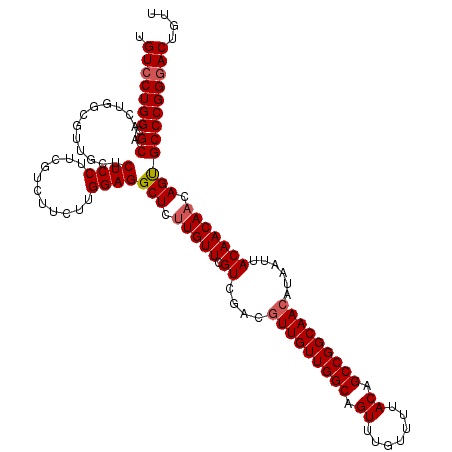
| Location | 17,261,542 – 17,261,662 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -43.06 |
| Consensus MFE | -36.80 |
| Energy contribution | -37.24 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.998181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17261542 120 - 23771897 UGUCCUGGCGCAACUGGCGUUGCUCUCCUUCGUCUUCUUGGAGGCUCUUGUUCGUCGACGUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGUGCCCGGGACUGUU .(((((((.(((.(((..((((....((((((......))))))...............(((((((((.((.......)).))))))))).......))))..))))))))))))).... ( -45.00) >DroSec_CAF1 145110 120 - 1 UGUCCUGGCGCAACUGGCGUUGCUCUCCUUCGUCUUCUUGGAGGCUCUUGUUCGUCGACGUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGUGCCCGGGACUGUU .(((((((.(((.(((..((((....((((((......))))))...............(((((((((.((.......)).))))))))).......))))..))))))))))))).... ( -45.00) >DroSim_CAF1 140011 120 - 1 UGUCCUGGCGCAACUGGCGUUGCUCUCCUUCGUCUUCUUGGAGGCUCUUGUUCGUCGACGUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGUGCCCGGGACUGUU .(((((((.(((.(((..((((....((((((......))))))...............(((((((((.((.......)).))))))))).......))))..))))))))))))).... ( -45.00) >DroEre_CAF1 148228 118 - 1 UGUACUGGCGCAACGGCAG--CAUCUCCUUCGUCUUCUUGGAGGCUCCUGUUCGUCCUCGUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGUGCCCGGGACUCUU .(((((((((.(((((.((--(..((((...........))))))).))))))))....(((((((((.((.......)).))))))))).............))))))........... ( -37.10) >DroYak_CAF1 152510 118 - 1 UGUCCUGGCGCAACGGCAG--GCGCUCCUUCGUCUUCUUGGAGGCUCUUGUUCGUCGUCCUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGCGCCCGGGACUGUU .((((((((((.(((((..--(((..((((((......))))))....)))..)))))..((((((((.((.......)).))))))))................))).))))))).... ( -43.20) >consensus UGUCCUGGCGCAACUGGCGUUGCUCUCCUUCGUCUUCUUGGAGGCUCUUGUUCGUCGACGUUGUUGGCAGUUUGUUUUACAGCCGGCAACAUAAUUACAACAACAGUGCCCGGGACUGUU .(((((((.((.............((((...........))))(((.(((((.((....(((((((((.((.......)).)))))))))......))))))).)))))))))))).... (-36.80 = -37.24 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:33 2006