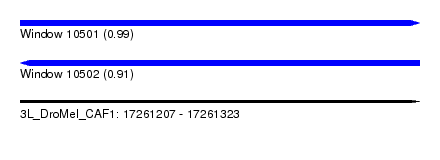
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,261,207 – 17,261,323 |
| Length | 116 |
| Max. P | 0.994594 |

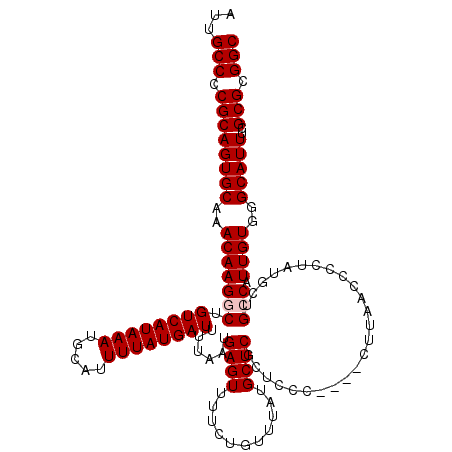
| Location | 17,261,207 – 17,261,323 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -31.22 |
| Energy contribution | -31.22 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17261207 116 + 23771897 GCCACGCGAAAUGCCCACAAUGGCAGCAUAGGGGUUAAG----GGGAGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.((((...((((......))))(((..((((((...----......(((.............))).......(((((((.....))))))).))))))...)))..)))).)))... ( -30.72) >DroSec_CAF1 144809 116 + 1 GCCGCGCGAAAUGCCCACAAUGGCAGCAUAGGGGUUAAG----GGGAGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.((((...((((......))))(((..((((((...----......(((.............))).......(((((((.....))))))).))))))...)))..)))).)))... ( -32.82) >DroSim_CAF1 139700 116 + 1 GCCGCGCGAAAUGCCCACAAUGGCAGCAUAGGGGUUAAG----GGGAGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.((((...((((......))))(((..((((((...----......(((.............))).......(((((((.....))))))).))))))...)))..)))).)))... ( -32.82) >DroEre_CAF1 147871 120 + 1 GCCGCGCGAAAUGCCCACAAUGCCAGCAUAGGGGUUAAGGUGGGGGGGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.(((......(((((...(((........)))....)))))((.(((((((......((....)).......(((((((.....)))))))......))))))).))))).)))... ( -37.60) >DroYak_CAF1 152148 120 + 1 GCCGCGCGAAAUGCCCACAAUGCCAACAUAGGGGUUAAAGGGGGGGUGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.(((......(((.....(((........))).....))).((((((((((......((....)).......(((((((.....)))))))......))))))))))))).)))... ( -38.20) >consensus GCCGCGCGAAAUGCCCACAAUGGCAGCAUAGGGGUUAAG____GGGAGCGAGCAUAAACAGAAAACUCAUUAAAAUCAUAAAAUGCAUUUAUGACAGCCUUGUUUGCACUGCGGGGCAAU (((.(((.....((((.(.(((....))).))))).........((.(((((((......((....)).......(((((((.....)))))))......))))))).))))).)))... (-31.22 = -31.22 + 0.00)

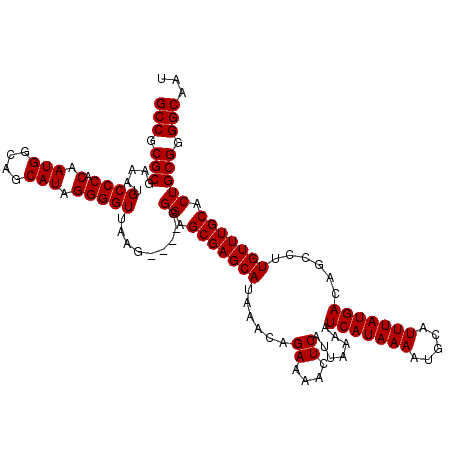

| Location | 17,261,207 – 17,261,323 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.79 |
| Mean single sequence MFE | -31.74 |
| Consensus MFE | -28.22 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17261207 116 - 23771897 AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCUCCC----CUUAACCCCUAUGCUGCCAUUGUGGGCAUUUCGCGUGGC ...(((.((((((((..(((((((.((((((((.....))))))))......((((...........))))......----...............))).))))..)))))..))).))) ( -31.30) >DroSec_CAF1 144809 116 - 1 AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCUCCC----CUUAACCCCUAUGCUGCCAUUGUGGGCAUUUCGCGCGGC ...(((.((((((((..(((((((.((((((((.....))))))))......((((...........))))......----...............))).))))..)))))..))).))) ( -31.30) >DroSim_CAF1 139700 116 - 1 AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCUCCC----CUUAACCCCUAUGCUGCCAUUGUGGGCAUUUCGCGCGGC ...(((.((((((((..(((((((.((((((((.....))))))))......((((...........))))......----...............))).))))..)))))..))).))) ( -31.30) >DroEre_CAF1 147871 120 - 1 AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCCCCCCCACCUUAACCCCUAUGCUGGCAUUGUGGGCAUUUCGCGCGGC ...(((.((((((((......(((.((((((((.....))))))))......((((...........)))))))...((((.......((......))....)))))))))..))).))) ( -31.20) >DroYak_CAF1 152148 120 - 1 AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCACCCCCCCUUUAACCCCUAUGUUGGCAUUGUGGGCAUUUCGCGCGGC ...((((((((((((.(((((((..((((((((.....))))))))......((((...........)))).................))).)))).)))))))))((.....))..))) ( -33.60) >consensus AUUGCCCCGCAGUGCAAACAAGGCUGUCAUAAAUGCAUUUUAUGAUUUUAAUGAGUUUUCUGUUUAUGCUCGCUCCC____CUUAACCCCUAUGCUGCCAUUGUGGGCAUUUCGCGCGGC ...(((.((((((((..(((((((.((((((((.....))))))))......((((...........)))).........................))).))))..)))))..))).))) (-28.22 = -28.62 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:31 2006