
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,260,349 – 17,260,951 |
| Length | 602 |
| Max. P | 0.999989 |

| Location | 17,260,349 – 17,260,464 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -42.72 |
| Energy contribution | -42.12 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.79 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.53 |
| SVM RNA-class probability | 0.999989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260349 115 + 23771897 AGGCAGUGCCAAAUAAAAGUGGACGGAUGGGCG-----UGGCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGU ..((..((((..((((((((((((((((.(((.-----..))).((((.....))))...))))).)))))))))))....((((..((((.......))))..))))))))...))... ( -44.70) >DroSec_CAF1 143954 115 + 1 AGGCAGUGCCAAAUAAAAGUGGCCGGAUGGGCG-----UGGCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGU ..((..((((..((((((((((.(((((.(((.-----..))).((((.....))))...)))))..))))))))))....((((..((((.......))))..))))))))...))... ( -42.00) >DroSim_CAF1 138838 115 + 1 AGGCAGUGCCAAAUAAAAGUGGCCGGAUGGGCG-----UGGCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAGUGCGGGUGAAACGGCAACAGCAGU ..((..((((..((((((((((.(((((.(((.-----..))).((((.....))))...)))))..))))))))))....((((..((((.......))))..))))))))...))... ( -42.00) >DroEre_CAF1 147033 115 + 1 AGGCAGUGCCAAAUAGAAGUGGCCGGAUGGGCG-----UGGCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGU ..((..((((..((((((((((.(((((.(((.-----..))).((((.....))))...)))))..))))))))))....((((..((((.......))))..))))))))...))... ( -42.10) >DroYak_CAF1 151248 120 + 1 AGGCAGUGCCAAAUGGAAGUGGCAGGGUGGGCGUGGCGUGGCCAGGUGGGAUGCACCAUGAUUCGAUACAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGU ..((..((((..(..((((((......(((((((((.((..((.....))..)).))))).))))...))))))..)....((((..((((.......))))..))))))))...))... ( -41.70) >consensus AGGCAGUGCCAAAUAAAAGUGGCCGGAUGGGCG_____UGGCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGU ..((..((((..(((((((((..(((((.(((........))).((((.....))))...)))))...)))))))))....((((..((((.......))))..))))))))...))... (-42.72 = -42.12 + -0.60)



| Location | 17,260,349 – 17,260,464 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.21 |
| Mean single sequence MFE | -27.66 |
| Consensus MFE | -24.22 |
| Energy contribution | -25.26 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260349 115 - 23771897 ACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGCCA-----CGCCCAUCCGUCCACUUUUAUUUGGCACUGCCU ...((.((.(((((((..((((.......))))..))))........(((((((..((...((((.((...((....))...-----.))))))...))....)))))))))))).)).. ( -26.40) >DroSec_CAF1 143954 115 - 1 ACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGCCA-----CGCCCAUCCGGCCACUUUUAUUUGGCACUGCCU ...((.((.(((((((..((((.......))))..))))........(((((((.........((((.....))))(((((.-----.........)))))..)))))))))))).)).. ( -29.90) >DroSim_CAF1 138838 115 - 1 ACUGCUGUUGCCGUUUCACCCGCACUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGCCA-----CGCCCAUCCGGCCACUUUUAUUUGGCACUGCCU ...((.((.(((((((..((((.......))))..))))........(((((((.........((((.....))))(((((.-----.........)))))..)))))))))))).)).. ( -29.90) >DroEre_CAF1 147033 115 - 1 ACUGCUGUUGCCGCUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGCCA-----CGCCCAUCCGGCCACUUCUAUUUGGCACUGCCU ...((.((.(((......((((.......))))..................((((.(((....((((.....))))(((((.-----.........))))).))).))))))))).)).. ( -25.40) >DroYak_CAF1 151248 120 - 1 ACUGCUGUUGCCGCUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGUAUCGAAUCAUGGUGCAUCCCACCUGGCCACGCCACGCCCACCCUGCCACUUCCAUUUGGCACUGCCU ...((.((.(((((.......)).......((((...............((((((((.....))))))))......((((...)))).))))..................))))).)).. ( -26.70) >consensus ACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGCCA_____CGCCCAUCCGGCCACUUUUAUUUGGCACUGCCU ...((.((.(((((((..((((.......))))..))))........(((((((.........((((.....))))(((((...............)))))..)))))))))))).)).. (-24.22 = -25.26 + 1.04)


| Location | 17,260,384 – 17,260,499 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -38.14 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260384 115 + 23771897 GCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAAU ....((((.....)))).((((((((((((...((((....((((..((((.......))))..))))(....).)))).))))))((((((....)))))).).))))-----)..... ( -38.20) >DroSec_CAF1 143989 112 + 1 GCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAA---CAAUC-----AAAACU (((.((((.....))))((((......))))..........((((..((((.......))))..)))))))......((((..(((((((((....)))))---)))).-----..)))) ( -39.30) >DroSim_CAF1 138873 115 + 1 GCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAGUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCGAACUGCAAUC-----AAAAAU ....((((.....)))).((((((((((((...((((....((((..((((.......))))..))))(....).)))).))))))((((((....)))))).).))))-----)..... ( -37.90) >DroEre_CAF1 147068 115 + 1 GCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAAC ....((((.....)))).((((((((((((...((((....((((..((((.......))))..))))(....).)))).))))))((((((....)))))).).))))-----)..... ( -38.20) >DroYak_CAF1 151288 120 + 1 GCCAGGUGGGAUGCACCAUGAUUCGAUACAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUCAAAUCAAAAAU ....(((..((((((....((((((((..............((((..((((.......))))..))))(....)....))))))))((((((....))))))))).)))..)))...... ( -37.10) >consensus GCCAGGUGGGAUGCACCAUGAUUCGAUUCAUUUUUGUAAUUGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC_____AAAAAU ....((((.....))))..(((((((((.....((((..((((((..((((.......))))..))))))..)))).)))))))))((((((....)))))).................. (-34.40 = -34.40 + -0.00)


| Location | 17,260,384 – 17,260,499 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -28.81 |
| Consensus MFE | -24.18 |
| Energy contribution | -24.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260384 115 - 23771897 AUUUUU-----GAUUGCAGUUUGCCCUGGCAAACAAUUCAACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGC ...(((-----((((.((((((((....))))))....((((....))))..((((..((((.......))))..))))...........)))))))))(((.((((.....))))))). ( -28.70) >DroSec_CAF1 143989 112 - 1 AGUUUU-----GAUUG---UUUGCCCUGGCAAACAAUUCAACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGC (((.((-----(((((---(((((....)))))))).))))..)))...(((((((..((((.......))))..))))..........((((......))))((((.....)))).))) ( -31.20) >DroSim_CAF1 138873 115 - 1 AUUUUU-----GAUUGCAGUUCGCCCUGGCAAACAAUUCAACUGCUGUUGCCGUUUCACCCGCACUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGC ...(((-----((((.(((((.((....)).)))....((((....))))..((((..((((.......))))..))))...........)))))))))(((.((((.....))))))). ( -24.60) >DroEre_CAF1 147068 115 - 1 GUUUUU-----GAUUGCAGUUUGCCCUGGCAAACAAUUCAACUGCUGUUGCCGCUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGC ((((((-----(((((...(((((((.(((((.((..........)))))))((.......)).......))))))).))))..)))))))........(((.((((.....))))))). ( -28.30) >DroYak_CAF1 151288 120 - 1 AUUUUUGAUUUGAUUGCAGUUUGCCCUGGCAAACAAUUCAACUGCUGUUGCCGCUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGUAUCGAAUCAUGGUGCAUCCCACCUGGC .....(((((((((.(((.(((((((.(((((.((..........)))))))((.......)).......))))))).............)))))))))))).((((.....)))).... ( -31.24) >consensus AUUUUU_____GAUUGCAGUUUGCCCUGGCAAACAAUUCAACUGCUGUUGCCGUUUCACCCGCAUUUUCCGGGCAAAACAAUUACAAAAAUGAAUCGAAUCAUGGUGCAUCCCACCUGGC ((((((.....(((((...(((((((.(((((.((..........)))))))((.......)).......))))))).)))))..))))))........(((.((((.....))))))). (-24.18 = -24.18 + -0.00)


| Location | 17,260,424 – 17,260,539 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.36 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -29.14 |
| Energy contribution | -28.82 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.62 |
| SVM RNA-class probability | 0.999453 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260424 115 + 23771897 UGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAAUGCGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUU .((((..((((.......))))..)))).(((((((..((((((((((((((....))))))((((...-----.....))))...............))))))))..))).)))).... ( -34.30) >DroSec_CAF1 144029 112 + 1 UGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAA---CAAUC-----AAAACUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUU .((((..((((.......))))..))))(....)..(((((..(((((((((....)))))---)))).-----..)))))...(((((..(((((.........))))).))))).... ( -35.50) >DroSim_CAF1 138913 115 + 1 UGUUUUGCCCGGAAAGUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCGAACUGCAAUC-----AAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUU .((((..((((.......))))..)))).(((((((..((((((((.(((((....)))))((((..((-----(....)))..))))..........))))))))..))).)))).... ( -33.50) >DroEre_CAF1 147108 115 + 1 UGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAACGGGAGUAGAAAAAAUAGAAGUUUAAUGUUUGUUUGCGCUU .((((..((((.......))))..))))(((((..(((((...))))))))))((.((((((.(((...-----....((....)).....((((....))))..)))..)))))).)). ( -33.60) >DroYak_CAF1 151328 120 + 1 UGUUUUGCCCGGAAAAUGCGGGUGAAGCGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUCAAAUCAAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUU .((((..((((.......))))..)))).(((((((..((((((((.(((((....)))))((((..(((.........)))..))))..........))))))))..))).)))).... ( -33.00) >consensus UGUUUUGCCCGGAAAAUGCGGGUGAAACGGCAACAGCAGUUGAAUUGUUUGCCAGGGCAAACUGCAAUC_____AAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUU .((((..((((.......))))..)))).(((((((..((((((((((((((....))))))................((....))............))))))))..))).)))).... (-29.14 = -28.82 + -0.32)



| Location | 17,260,464 – 17,260,577 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.40 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260464 113 + 23771897 UGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAAUGCGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUUCAAUUCAAUG--UUUGUUUGUCUGCCGGAAGGCAGUUGCU ((((((((((((....)))))).......-----..(((((((((((((......(....)......))))))))))))).))))))...--...((....(((((....)))))..)). ( -31.50) >DroSec_CAF1 144069 112 + 1 UGAAUUGUUUGCCAGGGCAAA---CAAUC-----AAAACUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUUCAAUUCAAUGUGUUUGUUUGCCUGCCGCAAGGCAGUUGCU (((.((((((((....)))))---)))))-----)((((...((((.((((....(.((((.....)))).).....)))).)))).....))))....(((((((....)))))..)). ( -31.00) >DroSim_CAF1 138953 115 + 1 UGAAUUGUUUGCCAGGGCGAACUGCAAUC-----AAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUUCAAUUCAAUGUGUUUGUUUGCCUGCCGCAAGGCAGUUGCU ......((((((....)))))).((((.(-----(((.....((((.((((....(.((((.....)))).).....)))).))))......)))).))))(((((....)))))..... ( -31.40) >DroEre_CAF1 147148 113 + 1 UGAAUUGUUUGCCAGGGCAAACUGCAAUC-----AAAAACGGGAGUAGAAAAAAUAGAAGUUUAAUGUUUGUUUGCGCUUCAAUUCAAUG--UUUGUUUGUCUGCCGCAAGGCAGUUGUU ......((((((....)))))).((((.(-----(((.((....))..........(((((.(((.......))).))))).........--)))).))))(((((....)))))..... ( -29.30) >DroYak_CAF1 151368 118 + 1 UGAAUUGUUUGCCAGGGCAAACUGCAAUCAAAUCAAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUUCAAUUCAAUG--UUUGUUUGUCUGCCGCAAGGCAGUUGUU ......((((((....)))))).((((.((((.((((.....((((.((((....(.((((.....)))).).....)))).))))....--)))))))).(((((....))))))))). ( -29.50) >consensus UGAAUUGUUUGCCAGGGCAAACUGCAAUC_____AAAAAUGGGAGUAGAAAAAAUAGAAAUUUAAUGUUUGUUUGCGUUUCAAUUCAAUG__UUUGUUUGUCUGCCGCAAGGCAGUUGCU ((((((((((((....))))))..................................(((((.(((.......))).)))))))))))..............(((((....)))))..... (-25.68 = -25.40 + -0.28)

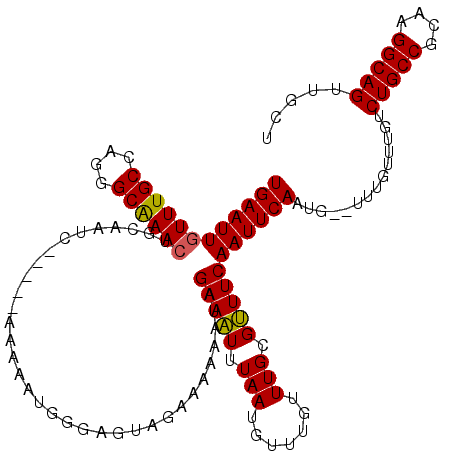
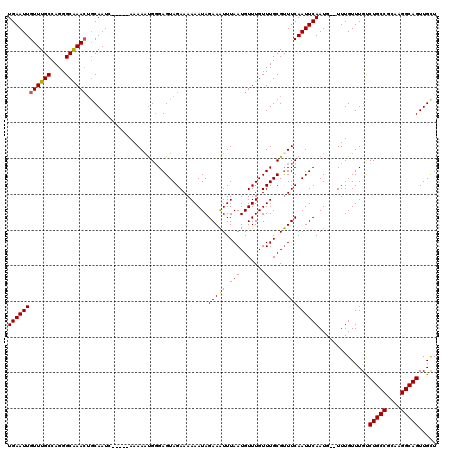
| Location | 17,260,464 – 17,260,577 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -23.49 |
| Consensus MFE | -17.74 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.924586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260464 113 - 23771897 AGCAACUGCCUUCCGGCAGACAAACAAA--CAUUGAAUUGAAACGCAAACAAACAUUAAAUUUCUAUUUUUUCUACUCGCAUUUUU-----GAUUGCAGUUUGCCCUGGCAAACAAUUCA .....(((((....))))).........--...((((((.....((((.((((..............................)))-----).)))).((((((....)))))))))))) ( -23.71) >DroSec_CAF1 144069 112 - 1 AGCAACUGCCUUGCGGCAGGCAAACAAACACAUUGAAUUGAAACGCAAACAAACAUUAAAUUUCUAUUUUUUCUACUCCCAGUUUU-----GAUUG---UUUGCCCUGGCAAACAAUUCA .((..(((((....)))))))............(((((..((((.............((((....))))............)))).-----.)(((---(((((....)))))))))))) ( -22.81) >DroSim_CAF1 138953 115 - 1 AGCAACUGCCUUGCGGCAGGCAAACAAACACAUUGAAUUGAAACGCAAACAAACAUUAAAUUUCUAUUUUUUCUACUCCCAUUUUU-----GAUUGCAGUUCGCCCUGGCAAACAAUUCA .((..(((((....)))))))............(((((((....((((.((((..............................)))-----).)))).....((....))...))))))) ( -21.81) >DroEre_CAF1 147148 113 - 1 AACAACUGCCUUGCGGCAGACAAACAAA--CAUUGAAUUGAAGCGCAAACAAACAUUAAACUUCUAUUUUUUCUACUCCCGUUUUU-----GAUUGCAGUUUGCCCUGGCAAACAAUUCA .....(((((....))))).........--...((((((.....((((.((((..............................)))-----).)))).((((((....)))))))))))) ( -23.71) >DroYak_CAF1 151368 118 - 1 AACAACUGCCUUGCGGCAGACAAACAAA--CAUUGAAUUGAAACGCAAACAAACAUUAAAUUUCUAUUUUUUCUACUCCCAUUUUUGAUUUGAUUGCAGUUUGCCCUGGCAAACAAUUCA .....(((((....))))).........--...((((((.....((((.(((..((((((.......................))))))))).)))).((((((....)))))))))))) ( -25.40) >consensus AGCAACUGCCUUGCGGCAGACAAACAAA__CAUUGAAUUGAAACGCAAACAAACAUUAAAUUUCUAUUUUUUCUACUCCCAUUUUU_____GAUUGCAGUUUGCCCUGGCAAACAAUUCA .....(((((....)))))..............(((((((....((((.............................................))))..(((((....)))))))))))) (-17.74 = -18.14 + 0.40)



| Location | 17,260,539 – 17,260,655 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -23.32 |
| Energy contribution | -23.56 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260539 116 + 23771897 CAAUUCAAUG--UUUGUUUGUCUGCCGGAAGGCAGUUGCUGCA--AUUGUUGUGGUUGUGCUUGUCACACGUGCUGGGCAUUGAUUUAAGCGAUUAUUUUAUUUGAUUUCUAAUUAAAAA (((((((((.--.((((....(((((....))))).....)))--)..)))).)))))((((((.((....)).))))))((((((..((.(((((.......))))).))))))))... ( -27.40) >DroSec_CAF1 144141 112 + 1 CAAUUCAAUGUGUUUGUUUGCCUGCCGCAAGGCAGUUGCUGCA--AUUGUUGUGGUU------GUCACUCGUGCUGGGCAUUGAUUUAAGAGAUUAUUUUAUUUGAUUUCUAAUUAAAAA ....(((((((........(((((((....)))))..)).(((--......(((...------..)))...)))...)))))))....((((((((.......))))))))......... ( -29.00) >DroSim_CAF1 139028 118 + 1 CAAUUCAAUGUGUUUGUUUGCCUGCCGCAAGGCAGUUGCUACA--AUUGUUGUGGUUGUGCCUGUCACACGUGCUGGGCAUUGAUUUAAGCGAUUAUUUUAUUUGAUUUCUAAUUAAAAA ....((((.(((.(((((((.(((((....)))))..((((((--.....)))))).(((((((.((....)).))))))).....))))))).))).....)))).............. ( -29.80) >DroEre_CAF1 147223 116 + 1 CAAUUCAAUG--UUUGUUUGUCUGCCGCAAGGCAGUUGUUGCA--AUUGCUGUGGUUGUGCUUGUCACACGUGCUGGGCAUUGAUUUAAGCGAUUAUUUUAUUUGAUUUCUAAUUAAAAA ....((((..--...((....(((((....))))).....))(--((((((......(((((((.((....)).))))))).......))))))).......)))).............. ( -27.82) >DroYak_CAF1 151448 118 + 1 CAAUUCAAUG--UUUGUUUGUCUGCCGCAAGGCAGUUGUUGCACACUUGUUGUGGUUGUGCUUGUCACACGUGCCGGGCAUUGAUUUAAGCGAUUAUUUUAUUUGAUUUCUAAUUAAAAA ....((((((--((((..((((((((....))))).....(((((((......)).))))).......)))...))))))))))....((.(((((.......))))).))......... ( -29.90) >consensus CAAUUCAAUG__UUUGUUUGUCUGCCGCAAGGCAGUUGCUGCA__AUUGUUGUGGUUGUGCUUGUCACACGUGCUGGGCAUUGAUUUAAGCGAUUAUUUUAUUUGAUUUCUAAUUAAAAA ....((((......(((....(((((....))))).....)))....((..((((((((....((((...((((...))))))))....))))))))..)).)))).............. (-23.32 = -23.56 + 0.24)

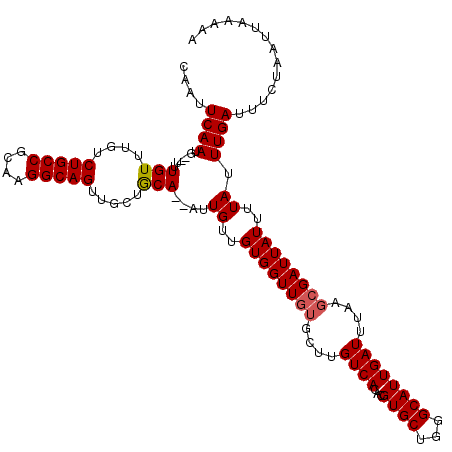

| Location | 17,260,539 – 17,260,655 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -16.20 |
| Energy contribution | -16.24 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260539 116 - 23771897 UUUUUAAUUAGAAAUCAAAUAAAAUAAUCGCUUAAAUCAAUGCCCAGCACGUGUGACAAGCACAACCACAACAAU--UGCAGCAACUGCCUUCCGGCAGACAAACAAA--CAUUGAAUUG ....................................((((((....(((..((((...........)))).....--))).....(((((....))))).........--)))))).... ( -18.50) >DroSec_CAF1 144141 112 - 1 UUUUUAAUUAGAAAUCAAAUAAAAUAAUCUCUUAAAUCAAUGCCCAGCACGAGUGAC------AACCACAACAAU--UGCAGCAACUGCCUUGCGGCAGGCAAACAAACACAUUGAAUUG ....................................((((((....(((...(((..------...)))......--))).((..(((((....))))))).........)))))).... ( -17.70) >DroSim_CAF1 139028 118 - 1 UUUUUAAUUAGAAAUCAAAUAAAAUAAUCGCUUAAAUCAAUGCCCAGCACGUGUGACAGGCACAACCACAACAAU--UGUAGCAACUGCCUUGCGGCAGGCAAACAAACACAUUGAAUUG ....................................((((((....((..((((.....))))....(((.....--))).))..(((((....)))))...........)))))).... ( -19.60) >DroEre_CAF1 147223 116 - 1 UUUUUAAUUAGAAAUCAAAUAAAAUAAUCGCUUAAAUCAAUGCCCAGCACGUGUGACAAGCACAACCACAGCAAU--UGCAACAACUGCCUUGCGGCAGACAAACAAA--CAUUGAAUUG ....................................((((((........((((.....)))).......((...--.)).....(((((....))))).........--)))))).... ( -18.90) >DroYak_CAF1 151448 118 - 1 UUUUUAAUUAGAAAUCAAAUAAAAUAAUCGCUUAAAUCAAUGCCCGGCACGUGUGACAAGCACAACCACAACAAGUGUGCAACAACUGCCUUGCGGCAGACAAACAAA--CAUUGAAUUG ....................................((((((....((((((((.....))))....((.....)))))).....(((((....))))).........--)))))).... ( -22.90) >consensus UUUUUAAUUAGAAAUCAAAUAAAAUAAUCGCUUAAAUCAAUGCCCAGCACGUGUGACAAGCACAACCACAACAAU__UGCAGCAACUGCCUUGCGGCAGACAAACAAA__CAUUGAAUUG ....................................((((((....(((..((((...........)))).......))).....(((((....)))))...........)))))).... (-16.20 = -16.24 + 0.04)


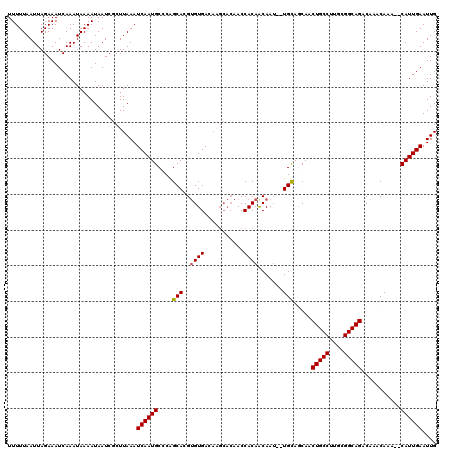
| Location | 17,260,655 – 17,260,768 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -19.42 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260655 113 - 23771897 GCAUAAUUUAUGCCAACGGACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAAAUGGAGGGCAUG----AAAAAAAAG-GCCGGCUGCUAUGA .(((..((((((((...(((((.(((.(((((((.((((........)))))))))))))))))))..(..((--......))..)))))))----))......(-((.....)))))). ( -26.70) >DroSec_CAF1 144253 113 - 1 GCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAUAUGGAGGGCAUG----AAAAAAAAA-GCCGACUGCUAUGA .((((.((((((((..(.((.(((((...(((((((((((((((..........))))))))))..)))))..--.))))))))..))))))----)).......-((.....)))))). ( -28.90) >DroSim_CAF1 139146 114 - 1 GCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAUAUGGAGGGCAUG----AAAAAAAAACGCCGACUGCUAUGA .((((.((((((((..(.((.(((((...(((((((((((((((..........))))))))))..)))))..--.))))))))..))))))----))........((.....)))))). ( -29.00) >DroEre_CAF1 147339 114 - 1 GCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAAUGCUCGCAUGUAGAGGGCAGG----AAAAAAGGCCAUGA--UGCUAUGA .((((..(((((((...(((((.((((.((.((((........)))).)).)))))))))...(((.....(((((.(.....).)))))))----).....)).)))))--...)))). ( -28.40) >DroYak_CAF1 151566 120 - 1 GCAUAAUUUAUGCCAAUGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACCAUACUCGUUUAUGGGGGGCAUGAACGAAAAAAAAAAAUGGUCUGGUAUGA .......(((((((..((((((.((((.((.((((........)))).)).))))))))))((.(((.......(((((((((.....)))))))))..........))).))))))))) ( -32.73) >consensus GCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA__CGCAUAUGGAGGGCAUG____AAAAAAAAA_GCCGACUGCUAUGA ........((((((...(((((.((((.((.((((........)))).)).)))))))))...((((..............)))).))))))............................ (-19.42 = -19.86 + 0.44)

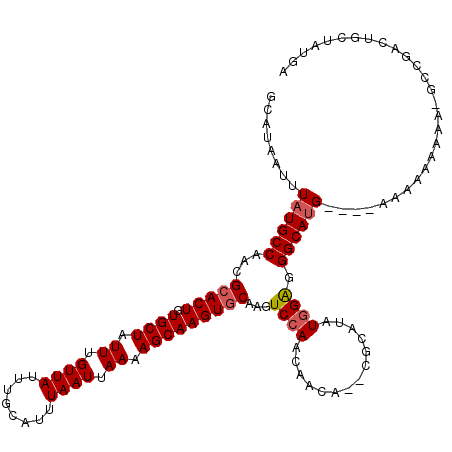
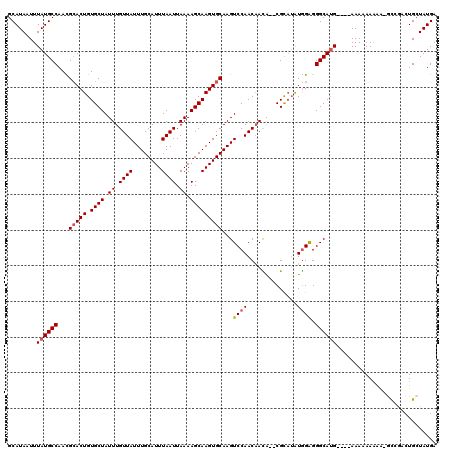
| Location | 17,260,690 – 17,260,800 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783624 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260690 110 + 23771897 UUGCG--UGUUGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUCCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU-CAGGGAAGGGUGUG------- (((((--(...((((((...((....)).))))))...))))))..........((((..(((.((((((((...))))))))...((.....))...-...)))...)))).------- ( -29.20) >DroSec_CAF1 144288 111 + 1 AUGCG--UGUUGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUUCCAGGGCAGGGUGAG------- .((((--(((((((....(((((.(((........))).)))))......)))))))).((((.((((((((...))))))))....))))..((......))))).......------- ( -28.90) >DroSim_CAF1 139182 111 + 1 AUGCG--UGUUGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUUCCAGGGCAGGGUGAG------- .((((--(((((((....(((((.(((........))).)))))......)))))))).((((.((((((((...))))))))....))))..((......))))).......------- ( -28.90) >DroEre_CAF1 147373 112 + 1 AUGCGAGCAUUGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU-CACAGUAGCUUGUG------- ..((((((((((((((....(((((((((.((..(((........)))..)).)))).))))).((((((((...))))))))..........)))..-.))))).)))))).------- ( -32.40) >DroYak_CAF1 151606 119 + 1 AAACGAGUAUGGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUGCAUUGGCAUAAAUUAUGCCAAAAAGGUACGACUGUU-CACGAAAGGGUGUGUGUGAGU ................(((..(((((((.(((....))).))))..........((((.((((.((((((((...))))))))....)))).......-..(....).)))))))..))) ( -29.90) >consensus AUGCG__UGUUGUUGGACUUGCACUUGCUUUUAAUUAAAUGCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU_CAGGGAAGGGUGUG_______ ...........((((.(((((((((((((.((..(((........)))..)).)))).))))).((((((((...))))))))..)))).)))).......................... (-22.70 = -22.90 + 0.20)


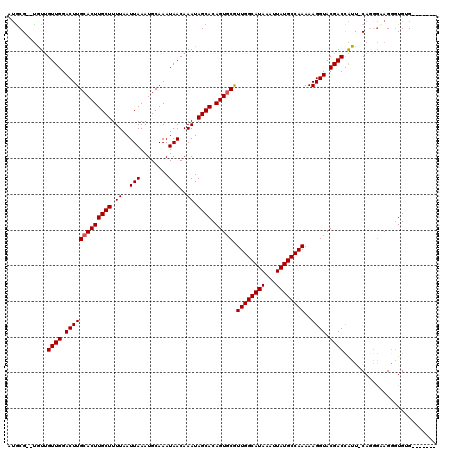
| Location | 17,260,690 – 17,260,800 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260690 110 - 23771897 -------CACACCCUUCCCUG-AAUGGUCGUACCUUUUUGGCAUAAUUUAUGCCAACGGACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAA -------.(((((.(((...)-)).))).))......((((((((...)))))))).(((((.(((.(((((((.((((........))))))))))))))))))).......--..... ( -26.70) >DroSec_CAF1 144288 111 - 1 -------CUCACCCUGCCCUGGAAUGGUCGUACCUUUUUGGCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAU -------.......(((..((...(((.(........((((((((...)))))))).(((((.((((.((.((((........)))).)).)))))))))..).)))....))--.))). ( -26.90) >DroSim_CAF1 139182 111 - 1 -------CUCACCCUGCCCUGGAAUGGUCGUACCUUUUUGGCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA--CGCAU -------.......(((..((...(((.(........((((((((...)))))))).(((((.((((.((.((((........)))).)).)))))))))..).)))....))--.))). ( -26.90) >DroEre_CAF1 147373 112 - 1 -------CACAAGCUACUGUG-AAUGGUCGUACCUUUUUGGCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAAUGCUCGCAU -------..........((((-(..((.....))...((((((((...)))))))).(((.(((...((((((.((((((.(((....))).))))))))))))...))).)))))))). ( -29.00) >DroYak_CAF1 151606 119 - 1 ACUCACACACACCCUUUCGUG-AACAGUCGUACCUUUUUGGCAUAAUUUAUGCCAAUGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACCAUACUCGUUU .......(((........)))-((((((.........((((((((...))))))))((((((.((((.((.((((........)))).)).))))))))))...........))).))). ( -24.80) >consensus _______CACACCCUGCCCUG_AAUGGUCGUACCUUUUUGGCAUAAUUUAUGCCAACGCACUGUGCUAUUUGUUAUUUGCAUUUAAUUAAAAGCAAGUGCAAGUCCAACAACA__CGCAU ........................(((.(........((((((((...)))))))).(((((.((((.((.((((........)))).)).)))))))))..).)))............. (-22.26 = -22.86 + 0.60)



| Location | 17,260,728 – 17,260,831 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.90 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260728 103 + 23771897 GCAAAUAACAAAUAGCACAGUCCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU-CAGGGAAGGGUGUG----------------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU (((.....(((...((((.((((.((((((((...))))))))...)).)).(((.((-(...))).))))))----------------)...))).((((.....)))).)))...... ( -30.80) >DroSec_CAF1 144326 104 + 1 GCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUUCCAGGGCAGGGUGAG----------------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU ..............((((.((((.((((((((...))))))))....))))..((((.....(((..(.(...----------------..).)..))).......))))))))...... ( -27.30) >DroSim_CAF1 139220 104 + 1 GCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUUCCAGGGCAGGGUGAG----------------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU ..............((((.((((.((((((((...))))))))....))))..((((.....(((..(.(...----------------..).)..))).......))))))))...... ( -27.30) >DroEre_CAF1 147413 103 + 1 GCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU-CACAGUAGCUUGUG----------------UGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU ..............((((.((((.((((((((...))))))))....))))..(((((-((((........))----------------)))).)))((((.....))))))))...... ( -30.10) >DroYak_CAF1 151646 119 + 1 GCAAAUAACAAAUAGCACAGUGCAUUGGCAUAAAUUAUGCCAAAAAGGUACGACUGUU-CACGAAAGGGUGUGUGUGAGUUUGAGUGUUUGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU .((((((.(((((.(((((((((.((((((((...))))))))....)))).......-..(....)....)))))..)))))..)))))).((...((((.....))))..))...... ( -30.70) >consensus GCAAAUAACAAAUAGCACAGUGCGUUGGCAUAAAUUAUGCCAAAAAGGUACGACCAUU_CAGGGAAGGGUGUG________________CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAU ..............((((.((((.((((((((...))))))))....))))..............................................((((.....))))))))...... (-21.70 = -21.90 + 0.20)



| Location | 17,260,800 – 17,260,911 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.02 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -24.56 |
| Energy contribution | -25.36 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260800 111 + 23771897 ---------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGU ---------.(((((((((((.....))))(((........))).(((((...(((((......)))))..))))).....)))))))...(((((((((........)))))))))... ( -28.90) >DroSec_CAF1 144399 111 + 1 ---------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCAGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGU ---------.(((((((((((.....))))(((........))).(((((...(((((.....)).)))..))))).....)))))))...(((((((((........)))))))))... ( -27.10) >DroSim_CAF1 139293 111 + 1 ---------CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCAGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGU ---------.(((((((((((.....))))(((........))).(((((...(((((.....)).)))..))))).....)))))))...(((((((((........)))))))))... ( -27.10) >DroEre_CAF1 147485 110 + 1 ---------UGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCAGCC-ACAAACUUUUAUAACACUCGCUCAACAUUGGGUGUUGCCU ---------.((((((((.........((((((....))))))..(((((...(((((......)))))..))))))))-)...))))...(((((((((........)))))))))... ( -26.60) >DroYak_CAF1 151725 120 + 1 UUGAGUGUUUGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACCCAACAAACUUUUAUAACACUCGCUUACCAUUGGGUGUUGCGU ..(((((((.(((((............((((((..(((((....)))))....(((((......))))).....))))))...)))))....)))))))((.((((....))))..)).. ( -31.66) >consensus _________CGAGUUGGCCAUUAUUUAUGGGUGCAAUCAUUCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGU ..........(((((((((((.....))))(((........))).(((((...(((((......)))))..))))).....)))))))...(((((((((........)))))))))... (-24.56 = -25.36 + 0.80)

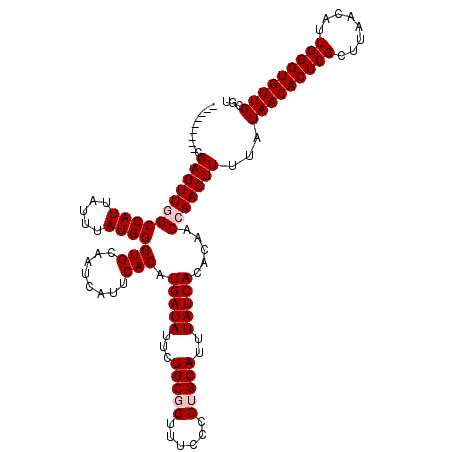
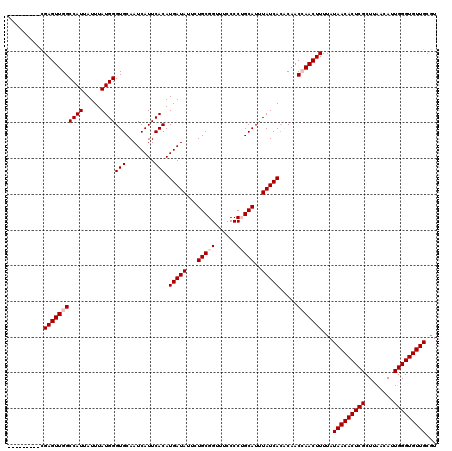
| Location | 17,260,831 – 17,260,951 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -25.06 |
| Consensus MFE | -20.84 |
| Energy contribution | -22.08 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260831 120 + 23771897 UCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGUUAUGAUUAAUGCUUUCCGCCACGAAGCGUUGCACUUAUAU .....(((((...(((((......)))))..)))))............(((((((.(.((((((....))))))..).))))))).(((((((((.......)))))))))......... ( -25.10) >DroSec_CAF1 144430 120 + 1 UCACAUGAUAUUCUGCGGUUUCCCCAGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGUUAUGAUUAAUGCUUUCCGCCACGAAGGGUUGCACUUAUAU .....(((((...(((((.....)).)))..)))))..(((((.........((((((((........))))))))((((((....))))))..............)))))......... ( -26.00) >DroSim_CAF1 139324 120 + 1 UCACAUGAUAUUCUGCGGUUUCCCCAGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGUUAUGAUUAAUGCUUUCCGCCGCGAAGGGUUGCACUUAUAU .....(((((...(((((.....)).)))..)))))..(((((.........((((((((........))))))))((((((....))))))..............)))))......... ( -26.00) >DroEre_CAF1 147516 117 + 1 UCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCAGCC-ACAAACUUUUAUAACACUCGCUCAACAUUGGGUGUUGCCUUAUGAUUAAUGCUUUCCGCCACGAACUGUUGCACUUAU-- .....(((((...(((((......)))))..)))))((.-(((.........((((((((........))))))))((.(((....))).))..............))).))......-- ( -22.00) >DroYak_CAF1 151765 120 + 1 UCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACCCAACAAACUUUUAUAACACUCGCUUACCAUUGGGUGUUGCGUUAUGAUUAAUGCUUUCCGCCACGAAGGAUUGCACUUAUAC .....(((((...(((((......)))))..)))))............(((((((.(.((((((....))))))..).)))))))....(((..(((........)))..)))....... ( -26.20) >consensus UCACAUGAUAUUCUGCGGUUUCCCCUGCAUUUAUCACACAACCAACUUUUAUAACACUCGCUUAACAUUGGGUGUUGCGUUAUGAUUAAUGCUUUCCGCCACGAAGGGUUGCACUUAUAU .....(((((...(((((......)))))..)))))..(((((.........((((((((........))))))))((((((....))))))..............)))))......... (-20.84 = -22.08 + 1.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:29 2006