| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,260,163 – 17,260,282 |
| Length | 119 |
| Max. P | 0.614166 |

| Location | 17,260,163 – 17,260,282 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -39.64 |
| Consensus MFE | -31.94 |
| Energy contribution | -31.82 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260163 119 + 23771897 AUAAAUUUAAGGGAAAAAAUGGCGA-AUGGCCGCAGGUCGGGUUGGCAUAAAAAAUGGCCCCAAGCCCACCGAUUUUCGAUACCCUCAACCCGCCGGCUCACUUGAGCGAUGGCCGGCGG .........((((.......(((..-...)))...(((.((((((((..........)))...))))))))...........))))....((((((((.(((......).)))))))))) ( -41.10) >DroSec_CAF1 143767 118 + 1 AUAAAUUUAAGGGAAAAAAUGGCGA-AUGGCCGCAGGUCGGGGUGGCAUAAAAAAUGGCCCAAAGCAC-CCGAUUUUCGAUCCCCUCAACCCGCUGGCUUACUUGAGCGAUGGCCGGCGG ..........((((......(((..-...)))(.(((((((((((((..........)))....)).)-))))))).)..))))......(((((((((............))))))))) ( -40.50) >DroSim_CAF1 138651 118 + 1 AUAAAUUUAAGGGAAAAAAUGGCGA-AUGGCCGCAGGUCGGGAUGGCAUAAAAAAUGGCCCCAAGCCC-CCGAUUCUCGAUCCCCUCAACCCGCUGGCUCACUUGAGCGAUGGCCGGCGG ..........((((......(((..-...)))(..(((((((..(((.................))))-))))))..)..))))......((((((((.(((......).)))))))))) ( -38.13) >DroEre_CAF1 146848 116 + 1 AUAAAUUCAAGGGAAAAAACGGCGAAAUGGCCGCAGGUCGGGAUGGCAUAAAAAAUGGCCUCAA-C---CCGAUUUUCGAUCCCCUCGUCCCGCUGGCUCACUUGAGCGAUGGCCGGCGG ..........((((.....((((......)))).((((((((..(((..........)))....-)---)))))))....))))......((((((((.(((......).)))))))))) ( -40.90) >DroYak_CAF1 151058 118 + 1 AUAAAUUCAAGGGAAAAAAUGGCGAAAUGGCCGCAGGUCGGAAUGAAAUAAAAAACGGCCUGAA-CUC-CCGAUUUUCGAUCCCCUCAUCCCGCUGGCGCACUUGAGCGAUGGCCGGCGG ..........((((......(((......))).(((((((...............)))))))..-.))-))...................((((((((.(((......).)))))))))) ( -37.56) >consensus AUAAAUUUAAGGGAAAAAAUGGCGA_AUGGCCGCAGGUCGGGAUGGCAUAAAAAAUGGCCCCAAGCCC_CCGAUUUUCGAUCCCCUCAACCCGCUGGCUCACUUGAGCGAUGGCCGGCGG .........((((......((((......))))..((((((((.(((..........)))..............))))))))))))....((((((((.(((......).)))))))))) (-31.94 = -31.82 + -0.12)

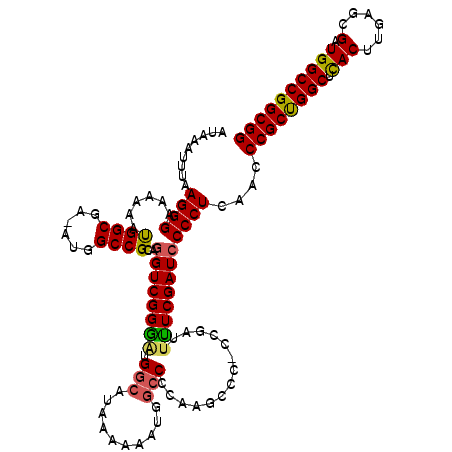
| Location | 17,260,163 – 17,260,282 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.18 |
| Mean single sequence MFE | -42.29 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.52 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17260163 119 - 23771897 CCGCCGGCCAUCGCUCAAGUGAGCCGGCGGGUUGAGGGUAUCGAAAAUCGGUGGGCUUGGGGCCAUUUUUUAUGCCAACCCGACCUGCGGCCAU-UCGCCAUUUUUUCCCUUAAAUUUAU ((((((((..((((....)))))))))))).(((((((....((((((..(..((.(((((..(((.....)))....)))))))..)(((...-..))))))))).)))))))...... ( -49.00) >DroSec_CAF1 143767 118 - 1 CCGCCGGCCAUCGCUCAAGUAAGCCAGCGGGUUGAGGGGAUCGAAAAUCGG-GUGCUUUGGGCCAUUUUUUAUGCCACCCCGACCUGCGGCCAU-UCGCCAUUUUUUCCCUUAAAUUUAU ((((.(((...(......)...))).)))).(((((((((.......((((-(.(.....(((.((.....))))).)))))).....(((...-..))).....)))))))))...... ( -36.70) >DroSim_CAF1 138651 118 - 1 CCGCCGGCCAUCGCUCAAGUGAGCCAGCGGGUUGAGGGGAUCGAGAAUCGG-GGGCUUGGGGCCAUUUUUUAUGCCAUCCCGACCUGCGGCCAU-UCGCCAUUUUUUCCCUUAAAUUUAU ((((.(((..((((....))))))).)))).(((((((((.........(.-(((.((((((.(((.....)))...))))))))).)(((...-..))).....)))))))))...... ( -43.30) >DroEre_CAF1 146848 116 - 1 CCGCCGGCCAUCGCUCAAGUGAGCCAGCGGGACGAGGGGAUCGAAAAUCGG---G-UUGAGGCCAUUUUUUAUGCCAUCCCGACCUGCGGCCAUUUCGCCGUUUUUUCCCUUGAAUUUAU ((((.(((..((((....))))))).))))..((((((((.......((((---(-....(((.((.....)))))..)))))...(((((......)))))...))))))))....... ( -44.90) >DroYak_CAF1 151058 118 - 1 CCGCCGGCCAUCGCUCAAGUGCGCCAGCGGGAUGAGGGGAUCGAAAAUCGG-GAG-UUCAGGCCGUUUUUUAUUUCAUUCCGACCUGCGGCCAUUUCGCCAUUUUUUCCCUUGAAUUUAU ((((.((((((.......))).))).))))..(.((((((..((((((.((-(((-....((((((....................))))))..))).)))))))))))))).)...... ( -37.55) >consensus CCGCCGGCCAUCGCUCAAGUGAGCCAGCGGGUUGAGGGGAUCGAAAAUCGG_GGGCUUGGGGCCAUUUUUUAUGCCAUCCCGACCUGCGGCCAU_UCGCCAUUUUUUCCCUUAAAUUUAU ((((.(((..((((....))))))).))))..((((((((.......((((.........(((.((.....)))))...)))).....(((......))).....))))))))....... (-31.44 = -31.52 + 0.08)

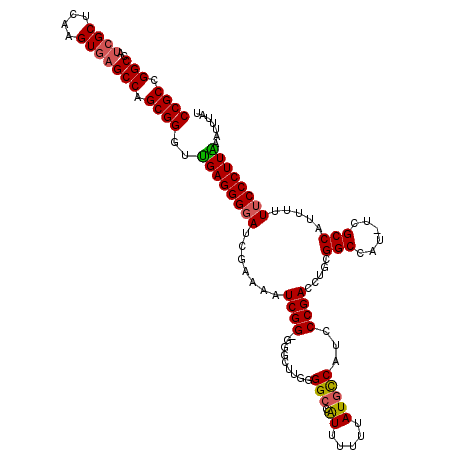
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:15 2006