
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,258,342 – 17,258,523 |
| Length | 181 |
| Max. P | 0.976184 |

| Location | 17,258,342 – 17,258,449 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -21.76 |
| Energy contribution | -21.52 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17258342 107 + 23771897 CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC------UGCAUCCGC------- ........(((((((..........)))))))(((.(((((..(((((...((((((.....(((....))).)))).))...))))).)))))....------)))......------- ( -26.30) >DroSec_CAF1 142008 108 + 1 CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUUCUCCGCAUUCGGCAUC------UGCAUCCGC------A .......(((((((((((...)))))......(((.(((((..((((((((..((((.....(((....))).))))..))).))))).)))))....------))).)))))------) ( -27.30) >DroSim_CAF1 136864 114 + 1 CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC------UGCAUCCGCAUACGCA ........(((((((..........)))))))((..(((((..(((((...((((((.....(((....))).)))).))...))))).)))))....------(((....)))...)). ( -28.10) >DroEre_CAF1 145099 107 + 1 CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAGAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGC-------------AUCUGCAUCCGCAGCCGCA ........(((((((..........)))))))((.........(((((...((((((.....(((....))).)))).))...)))))-------------..((((....))))..)). ( -25.70) >DroYak_CAF1 149239 120 + 1 CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAGAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUCUGCAUCUGCAUCCGCAUCCGCA .......((((((...................(((.(((((..(((((...((((((.....(((....))).)))).))...))))).)))))....)))...(((....))))))))) ( -31.50) >consensus CAUCCCUUGUGGAAAUUAUUUUAAUUUUUCACGCAACCGAAAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC______UGCAUCCGCA__CGCA ........(((((............(((((........)))))(((((...((((((.....(((....))).)))).))...)))))....................)))))....... (-21.76 = -21.52 + -0.24)



| Location | 17,258,342 – 17,258,449 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.37 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -26.60 |
| Energy contribution | -27.80 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17258342 107 - 23771897 -------GCGGAUGCA------GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG -------....(((((------...((((((.((((((.((..((((((((....))))....))))..))))))))..)))))))))))................((((....)))).. ( -32.50) >DroSec_CAF1 142008 108 - 1 U------GCGGAUGCA------GAUGCCGAAUGCGGAGAAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG .------....(((((------...((((((.(((((((.((.((((((((....))))....)))))).)))))))..)))))))))))................((((....)))).. ( -34.30) >DroSim_CAF1 136864 114 - 1 UGCGUAUGCGGAUGCA------GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG .((((((((....)))------...((((((.((((((.((..((((((((....))))....))))..))))))))..)))))))))))................((((....)))).. ( -33.70) >DroEre_CAF1 145099 107 - 1 UGCGGCUGCGGAUGCAGAU-------------GCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUCUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG .(((((((.(((.......-------------((((((.((..((((((((....))))....))))..))))))))))).)))))))..................((((....)))).. ( -29.61) >DroYak_CAF1 149239 120 - 1 UGCGGAUGCGGAUGCAGAUGCAGAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUCUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG ((.((((((....))).(((((...((((((.((((((.((..((((((((....))))....))))..))))))))..))))))))))).................))).))....... ( -35.50) >consensus UGCG__UGCGGAUGCA______GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUUUCGGUUGCGUGAAAAAUUAAAAUAAUUUCCACAAGGGAUG ...........(((((.........((((((.((((((.((..((((((((....))))....))))..))))))))..)))))))))))................((((....)))).. (-26.60 = -27.80 + 1.20)


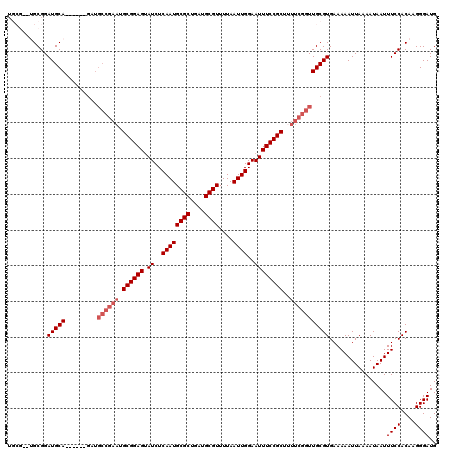
| Location | 17,258,382 – 17,258,483 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -20.35 |
| Energy contribution | -20.35 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17258382 101 + 23771897 AAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC------UGCAUCCGC------------AUAC-GCAUCCGCAUUUCGUAUCCGAAUCCGAAAG ...(((((...((((((.....(((....))).)))).))...))))).(((((....------(((....((------------....-))....))).((((....)))).))))).. ( -23.30) >DroSec_CAF1 142048 107 + 1 AAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUUCUCCGCAUUCGGCAUC------UGCAUCCGC------AUCCGCAUAC-GCAUCCGCAUUUCGUAUCCGAAUCCGAAAG ...((((((((..((((.....(((....))).))))..))).))))).(((((....------(((....))------)..((.((((-(.(........)))))).))...))))).. ( -25.50) >DroSim_CAF1 136904 113 + 1 AAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC------UGCAUCCGCAUACGCAUCCGCAUAU-GCAUCCGCAUUUCGUAUCCAAAUCCGAAAG ...(((((...((((((.....(((....))).)))).))...))))).(((((.((.------((.....(((((.((....)).)))-))....((.....))...)).))))))).. ( -26.50) >DroYak_CAF1 149279 120 + 1 AGAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUCUGCAUCUGCAUCCGCAUCCGCAUCCGCAUAUGGCAUCCGCAUAUCGCAUCCGAAUCCGAAAG ...(((((...((((((.....(((....))).)))).))...))))).(((((..(((((...(((....)))...)))...(((((((.......))))).))....))..))))).. ( -29.30) >consensus AAAGCGGAAAUUCCAAUUAAAACGCAUCAGCGCAUUGAGAUACUCCGCAUUCGGCAUC______UGCAUCCGC______AUCCGCAUAC_GCAUCCGCAUUUCGUAUCCGAAUCCGAAAG ...(((((...((((((.....(((....))).)))).))...))))).(((((...........((....)).................((....))...............))))).. (-20.35 = -20.35 + -0.00)



| Location | 17,258,382 – 17,258,483 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.02 |
| Mean single sequence MFE | -38.53 |
| Consensus MFE | -29.49 |
| Energy contribution | -29.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899461 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17258382 101 - 23771897 CUUUCGGAUUCGGAUACGAAAUGCGGAUGC-GUAU------------GCGGAUGCA------GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUU ..(((((.((((....)))).((((..(((-....------------)))..))))------....))))).((((((.((..((((((((....))))....))))..))))))))... ( -33.80) >DroSec_CAF1 142048 107 - 1 CUUUCGGAUUCGGAUACGAAAUGCGGAUGC-GUAUGCGGAU------GCGGAUGCA------GAUGCCGAAUGCGGAGAAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUU ..(((((.((((....)))).((((..(((-((......))------)))..))))------....))))).(((((((.((.((((((((....))))....)))))).)))))))... ( -38.10) >DroSim_CAF1 136904 113 - 1 CUUUCGGAUUUGGAUACGAAAUGCGGAUGC-AUAUGCGGAUGCGUAUGCGGAUGCA------GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUU ..(((((.((((....)))).((((..(((-((((((....)))))))))..))))------....))))).((((((.((..((((((((....))))....))))..))))))))... ( -42.40) >DroYak_CAF1 149279 120 - 1 CUUUCGGAUUCGGAUGCGAUAUGCGGAUGCCAUAUGCGGAUGCGGAUGCGGAUGCAGAUGCAGAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUCU .......((((((.((((...((((..(((.((.(((....))).)))))..))))..))))....))))))((((((.((..((((((((....))))....))))..))))))))... ( -39.80) >consensus CUUUCGGAUUCGGAUACGAAAUGCGGAUGC_AUAUGCGGAU______GCGGAUGCA______GAUGCCGAAUGCGGAGUAUCUCAAUGCGCUGAUGCGUUUUAAUUGGAAUUUCCGCUUU ..(((((.((((....)))).((((..(((.................)))..))))..........))))).((((((.((..((((((((....))))....))))..))))))))... (-29.49 = -29.55 + 0.06)

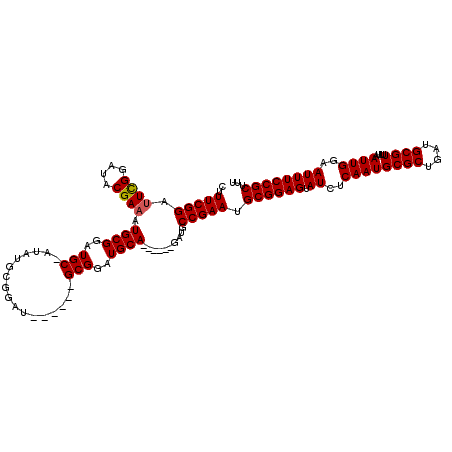

| Location | 17,258,422 – 17,258,523 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -23.42 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17258422 101 - 23771897 UUUGGAUUCGGAUCCUGACUCUGAUUCUGAUUCUGUUUCUCUUUCGGAUUCGGAUACGAAAUGCGGAUGC-GUAU------------GCGGAUGCA------GAUGCCGAAUGCGGAGUA ((((.((((((...(((..((((..(((((.((((.........)))).)))))......(((((....)-))))------------.))))..))------)...)))))).))))... ( -30.80) >DroSec_CAF1 142088 107 - 1 UUUGGAUUCGUAUCCUGACUCUGAUUCUGAUUCUGUUUCUCUUUCGGAUUCGGAUACGAAAUGCGGAUGC-GUAUGCGGAU------GCGGAUGCA------GAUGCCGAAUGCGGAGAA ((((.((((((((((....(((((.(((((.............))))).)))))......(((((....)-))))..))))------)))))).))------))..(((....))).... ( -34.52) >DroSim_CAF1 136944 113 - 1 UUUGGAUUCGGAUCCUGACUCUGAUUCUGAUUCUGUUUCUCUUUCGGAUUUGGAUACGAAAUGCGGAUGC-AUAUGCGGAUGCGUAUGCGGAUGCA------GAUGCCGAAUGCGGAGUA ((((.(((((((((.....((..(.(((((.............))))).)..)).......((((..(((-((((((....)))))))))..))))------))).)))))).))))... ( -38.82) >DroYak_CAF1 149319 114 - 1 UUUGGAUUCGGAUCCUGACUCUGAUUC------UGUUUCUCUUUCGGAUUCGGAUGCGAUAUGCGGAUGCCAUAUGCGGAUGCGGAUGCGGAUGCAGAUGCAGAUGCCGAAUGCGGAGUA ((((.((((((...(((..((((..((------(((.(((..((((.((..((((.((.....)).)).))..)).))))...))).)))))..))))..)))...)))))).))))... ( -37.70) >consensus UUUGGAUUCGGAUCCUGACUCUGAUUCUGAUUCUGUUUCUCUUUCGGAUUCGGAUACGAAAUGCGGAUGC_AUAUGCGGAU______GCGGAUGCA______GAUGCCGAAUGCGGAGUA ((((.(((((((((((((.(((((....((.......))....))))).))))........((((..(((.................)))..))))......))).)))))).))))... (-23.42 = -23.98 + 0.56)

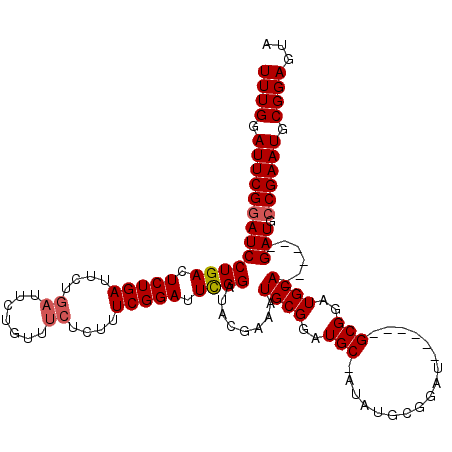
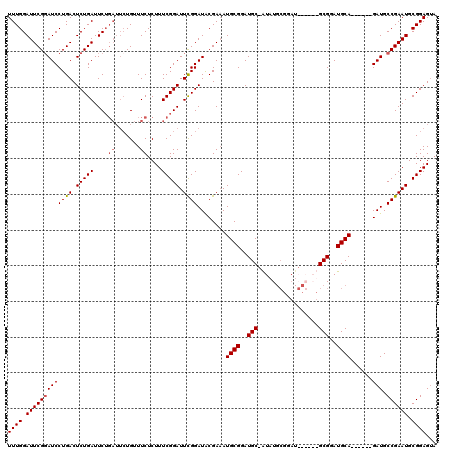
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:08 2006