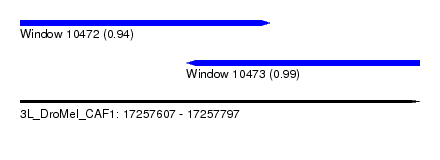
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,257,607 – 17,257,797 |
| Length | 190 |
| Max. P | 0.991596 |

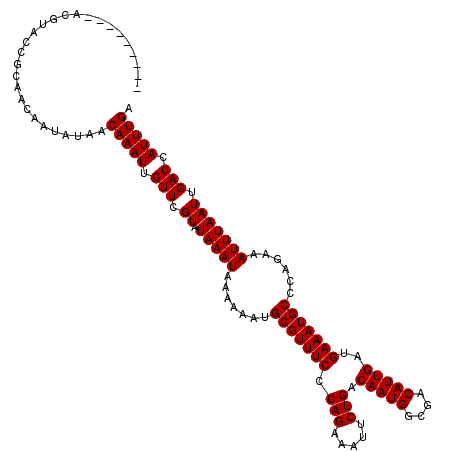
| Location | 17,257,607 – 17,257,726 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -33.86 |
| Energy contribution | -34.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17257607 119 + 23771897 GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCG-GAAAAAUCGGAGCCGGGGACGGAAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((...............(.-((....)).)..((((.((((....).))).)).))))))((((.......)))).............))))))).)......... ( -33.90) >DroSec_CAF1 141318 120 + 1 GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCGGGAAAAAUCGGAGCUGGGGACGGAAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((...............(((......)))...((((.((((....).))).)))).))))((((.......)))).............))))))).)......... ( -37.30) >DroSim_CAF1 136143 120 + 1 GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCGGGAAAAAUCGGAGCUGGGGACGGAAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((...............(((......)))...((((.((((....).))).)))).))))((((.......)))).............))))))).)......... ( -37.30) >DroEre_CAF1 144404 118 + 1 GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCG-GAA-AAUCGGAGCUGGGGACGGGAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((...............((-...-...))...((((.(((((...))))).)))).))))((((.......)))).............))))))).)......... ( -35.50) >DroYak_CAF1 148502 119 + 1 GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCG-GAAAAAUCGGAGCUGGGGACGGAAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((...............(.-((....)).)..((((.((((....).))).)))).))))((((.......)))).............))))))).)......... ( -36.20) >consensus GCGAUGCCCUAAGCCCACAUCAAACAAAAAGCG_GAAAAAUCGGAGCUGGGGACGGAAACUGUCGCCAGGGGGCGCCACAUCAAAUGGUCAAUUAAAUUUCUGGGCAUUUCAUCAAUGUC (.(((((((...((((..................((....))....((((.((((....).))).)))).))))((((.......)))).............))))))).)......... (-33.86 = -34.06 + 0.20)


| Location | 17,257,686 – 17,257,797 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -20.82 |
| Energy contribution | -20.82 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17257686 111 - 23771897 ---------ACGUACCGCAACAAUAUAACAAAUUGUUCGUUUAAAUAAAAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA ---------...................(((((.(((...((((((.......(((((((.(((.....))).(((((....)))))..)))))))......))))))..))).))))). ( -21.92) >DroSec_CAF1 141398 111 - 1 ---------ACGUACCGCAACAAUAUAACAAAUUGUUCGUAUAAAUAAAAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA ---------...................(((((.(((.((.(((((.......(((((((.(((.....))).(((((....)))))..)))))))......))))))).))).))))). ( -20.82) >DroSim_CAF1 136223 111 - 1 ---------ACGUACCGCAACAAUAUAACAAAUUGUUCGUAUAAAUAAAAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA ---------...................(((((.(((.((.(((((.......(((((((.(((.....))).(((((....)))))..)))))))......))))))).))).))))). ( -20.82) >DroEre_CAF1 144482 120 - 1 CCGCAACAUAGCUGCCGCAACAAUAUAACAAAUUGUUCGUAUAAAUAAGAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA ..(((.......))).............(((((.(((.((.(((((.......(((((((.(((.....))).(((((....)))))..)))))))......))))))).))).))))). ( -22.02) >DroYak_CAF1 148581 120 - 1 CUACAAAAUAGCUGCCGCAACAAUAUAACAAAUUGUUCGUAUAAAUAAAAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA .....((((..(((.((.((((((.......))))))))..............(((((((.(((.....))).(((((....)))))..))))))).)))..)))).............. ( -22.30) >consensus _________ACGUACCGCAACAAUAUAACAAAUUGUUCGUAUAAAUAAAAAAUGCGUUUCCCAGAAAUUCUGACAAUGGCGACAUUGAUGAAAUGCCCAGAAAUUUAAUUGACCAUUUGA ............................(((((.(((.((.(((((.......(((((((.(((.....))).(((((....)))))..)))))))......))))))).))).))))). (-20.82 = -20.82 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:04 2006