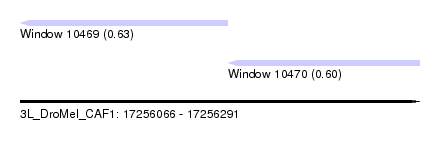
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,256,066 – 17,256,291 |
| Length | 225 |
| Max. P | 0.626960 |

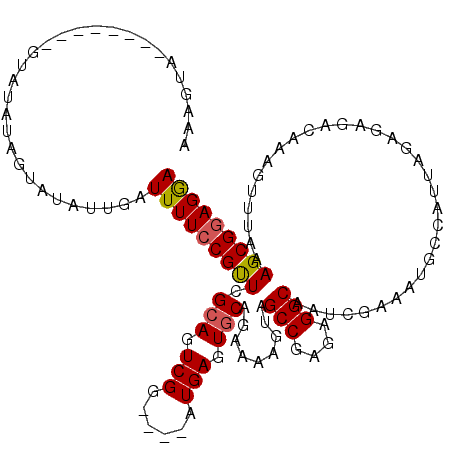
| Location | 17,256,066 – 17,256,183 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -24.62 |
| Consensus MFE | -20.20 |
| Energy contribution | -19.84 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17256066 117 - 23771897 ACAGGACCCAUGUCGAGCAUUGUCCAAAUACGAAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGCAGUAUUC---UCCGCCGCAUCCUUCACAUCUUCCA ...(((...((((.((((((((..((((((..(.(((((((.....))))))).)..))))))..))).)))..........(((.(.....---.).))).......))))))..))). ( -25.50) >DroSec_CAF1 139781 117 - 1 ACAGGACCCAUGUCGAGCAUUGUCCAAAUUCGAAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGCAGUAUCC---UCCGCCGCAUCCUCCGCAUCCUUCA ..((((.....((.(((.(((((((.........(((((((.....)))))))......(((((......)))))......)))))))...)---)).)).((.......)).))))... ( -23.10) >DroSim_CAF1 134617 117 - 1 ACAGGACCCAUGUCGAGCAUUGUCCAAAUACGAAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGAAGUAUCC---UCCGCCGCAUCCUCCACAUCUUCCA ...(((...((((.((((((((..((((((..(.(((((((.....))))))).)..))))))..))).))).........((((....)))---)...........)).))))..))). ( -24.30) >DroEre_CAF1 142906 117 - 1 ACAGGACCCAUGUCGAGCAUUGACCAAAUACGGAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGCAGUAUCC---UUCGCCGUAUCCUCCGCAUCCUCCG ...(((...((((.(((..........((((((.(((((((.....))))))).......((((((...((......))..)))))).....---....)))))).))).))))..))). ( -26.30) >DroYak_CAF1 146886 120 - 1 ACAGGACCCAUGUCGAGCAUUGACCAAAUACGAAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGCAUCCUCCACAUCCUCCGCAUCCUCCACAUCCUUUG ...(((...((((.(((...((...(((((..(.(((((((.....))))))).)..)))))((((...((......))..))))........))..))).))))..))).......... ( -23.90) >consensus ACAGGACCCAUGUCGAGCAUUGUCCAAAUACGAAAAGUAGCAAAAAGUUAUUUAUUAUAUUUGCCCGACUGCAAACACAAAGGGCAGUAUCC___UCCGCCGCAUCCUCCACAUCCUCCA ...(((...((((.((((((((..((((((..(.(((((((.....))))))).)..))))))..))).)))..........(((.............)))......)).))))..))). (-20.20 = -19.84 + -0.36)

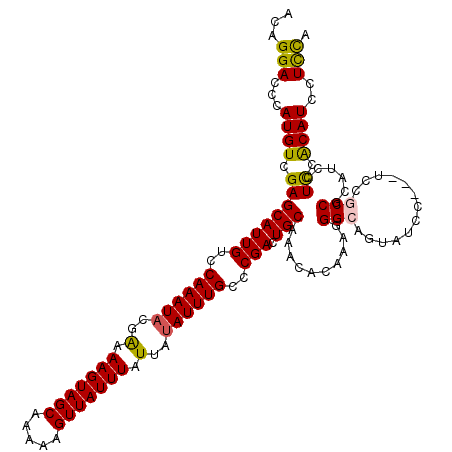
| Location | 17,256,183 – 17,256,291 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.14 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.58 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17256183 108 - 23771897 AAAGUA--------GUAUAUAGUAUAUUGAUUUUCCGUUCGCAUUCGG----AUGAGUGCAGAAAAGUAGCCGAGAGGCAAUCGAAAUGCCAUUAGAGAGACAAAGUUUAAAGCGGAGGA ....((--------(((((...))))))).(((((((((.((((((..----..)))))).........(((....)))................................))))))))) ( -24.30) >DroSec_CAF1 139898 112 - 1 AAAGUA--------GUAUAUAGUAUAUUGAUUUUCCGUUCGCAGUCGGACGGAUGAGUGCAGAAAAGUAGCCGAGAGGCUAUCGAAAUGCCAUUAGAGAGACAAAGUUUAAAGCGGAGGA ....((--------(((((...))))))).(((((((((.((.(((.....((((...(((.....((((((....)))))).....))))))).....)))...))....))))))))) ( -27.20) >DroSim_CAF1 134734 112 - 1 AAAGUA--------GUAUAUAGUAUAUUGAUUUUCCGUUCGCAGUCGGAUGGAUGAGUGCAGAAAAGUAGCCGAGAGGCAAUCGAAAUGCCAUUAGAGAGACAAAGUUUAAAGCGGAGGA ....((--------(((((...))))))).(((((((((.(((.(((......))).))).........(((....)))................................))))))))) ( -24.10) >DroEre_CAF1 143023 97 - 1 -------------------UAGUGUAUUGAUUUUCCGCUCGCAGUCGG----AUGAGUGCGGAAAAGUCGCCGAGAGGCAAUCGAAAUGCCAUUAGAGAGACAAAGUUUAAAGCGCAGAA -------------------..((((.((((((((((((.(.((.....----.)).).))))))))((((((....)))..((............))..))).....)))).)))).... ( -28.10) >DroYak_CAF1 147006 116 - 1 GAAGUAUAUAUAGUAUAUAUAAUGUAUUGAUUUUCCGUUCGGAAUCGG----AUGAGUGCAGAAAAGUAGCCGAGAGGCAAUCGAAAAGCCAUUAGAGAGACAAAGUUUAAAGCGGAGAA ..((((((((((.....))).)))))))..(((((((((.((..((((----.....(((......)))(((....)))..))))....)).......((((...))))..))))))))) ( -23.10) >consensus AAAGUA________GUAUAUAGUAUAUUGAUUUUCCGUUCGCAGUCGG____AUGAGUGCAGAAAAGUAGCCGAGAGGCAAUCGAAAUGCCAUUAGAGAGACAAAGUUUAAAGCGGAGGA ..............................(((((((((.(((.(((......))).))).........(((....)))................................))))))))) (-20.58 = -20.58 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:21:01 2006