
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,254,315 – 17,254,524 |
| Length | 209 |
| Max. P | 0.955798 |

| Location | 17,254,315 – 17,254,406 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -19.12 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.22 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17254315 91 + 23771897 UGCCACUUUGCCAGCUUUGACACUUUUUGUGGCUUACAGAAAAUUCCACAAAAAA-AAAAGAAAAUCAACAUAU--------------------ACGCAGAGCAAAAAAGAA .....((((....((((((.(..(((((((((..(.......)..))))))))).-....(.....).......--------------------..)))))))...)))).. ( -15.60) >DroSec_CAF1 138097 90 + 1 UGCCACUUUGCCAGCUUUGACACUUUUUGUGGCUUACAGAAAAUUCCACAAAAA--AAACGUAAAUCAACAUAU--------------------ACGCAGGGCAAAAAAGAA .....((((....((((((....(((((((((..(.......)..)))))))))--...((((.((....)).)--------------------)))))))))...)))).. ( -16.80) >DroSim_CAF1 132936 89 + 1 UGCCACUUUGCCAGCUUUGACACUUUUUGUGGCUUACAGAAAAUUCCACAAAA---AAAUGUAAAUCAACAUAU--------------------ACGUAGGGCAAAAAAGAA ......((((((...........(((((((((..(.......)..))))))))---).((((......))))..--------------------......))))))...... ( -16.60) >DroYak_CAF1 145068 108 + 1 UGCCACUUUGCCAGCCCUGACAGUUUUUGUGGCUUACAGCACAUGCCACAAAAAAAAAGAGCAAAUCAACAUAUUCUCGUUUAUGUAGCUUCAAACGUAGGGCAAA----AA ......((((((.((.((.....((((((((((...........))))))))))...)).))......(((((........)))))..............))))))----.. ( -27.50) >consensus UGCCACUUUGCCAGCUUUGACACUUUUUGUGGCUUACAGAAAAUUCCACAAAAA__AAAAGUAAAUCAACAUAU____________________ACGCAGGGCAAAAAAGAA .............((((((.(..(((((((((.............)))))))))......(........)..........................)))))))......... (-13.78 = -13.22 + -0.56)

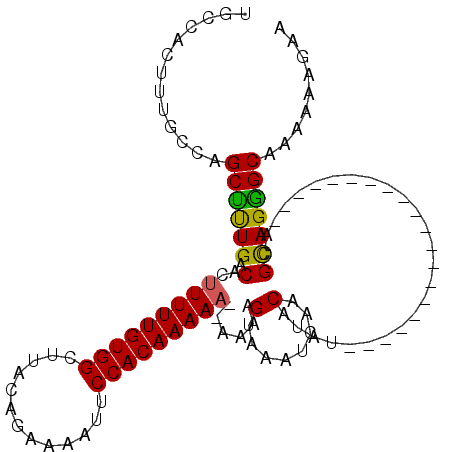

| Location | 17,254,406 – 17,254,524 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -20.06 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17254406 118 + 23771897 ACGCAUUACACAAACAUGACCGAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCCCUGGUGUUUUUUUCGUG--UGUCCUGUUGAUAUUU ............((((.((((((((...((((((.(....((((.(((((((((((((....)))))))))))))..))))......).)))))))))))..--.))).))))....... ( -22.40) >DroSec_CAF1 138187 120 + 1 ACGCAUUACACAAACAUGACCAAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCCCUGGUGUUUUUUUCGUGUGUGUCCUGUUGAUAUUU ..(((..((((..((((((..(((((......(((.....((((.(((((((((((((....)))))))))))))..))))....)))..)))))..))))))))))..)))........ ( -24.20) >DroSim_CAF1 133025 120 + 1 ACGCAUUACACAAACAUGACCGAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCCCUGGUGUUUUUUUCGUGUGUGUCCUGUUGAUAUUU ..(((..((((..((((((..(((((......(((.....((((.(((((((((((((....)))))))))))))..))))....)))..)))))..))))))))))..)))........ ( -24.60) >DroEre_CAF1 141341 118 + 1 ACGCAUUACACAAACAUGACCGAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCUCUGGUGUUUUUUUCGUU--UGUCCUGUUGAUAUUU ............((((.((((((((...((((((.(....((((.(((((((((((((....)))))))))))))..))))......).)))))))))))..--.))).))))....... ( -22.40) >DroYak_CAF1 145176 118 + 1 ACGCAUUACACAAACAUGACCAAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCCCUGGUGUUCUUUUCGUU--UGUCCUGUUGAUAUUU ............((((.(((..((((.(((((((.(....((((.(((((((((((((....)))))))))))))..))))......).)).)))))..)))--)))).))))....... ( -20.90) >consensus ACGCAUUACACAAACAUGACCGAAACCAAAGACAGCUUGUUAAUUGGAAAGUUUUAUGCAAUUAUAAAACUUUUUUCAUUAUUCCCUGGUGUUUUUUUCGUG__UGUCCUGUUGAUAUUU ............((((.(((....((.(((((((.(....((((.(((((((((((((....)))))))))))))..))))......).)))))))...))....))).))))....... (-20.06 = -20.26 + 0.20)



| Location | 17,254,406 – 17,254,524 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.32 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -21.22 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.772362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17254406 118 - 23771897 AAAUAUCAACAGGACA--CACGAAAAAAACACCAGGGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUCGGUCAUGUUUGUGUAAUGCGU ..((((.((((.(((.--..((((......((((((((.((((....((((((((((......))))))))))...))))........))))))))))))))).)))).))))....... ( -22.30) >DroSec_CAF1 138187 120 - 1 AAAUAUCAACAGGACACACACGAAAAAAACACCAGGGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUUGGUCAUGUUUGUGUAAUGCGU .........((..(((((.(((..(..((.((((((((.((((....((((((((((......))))))))))...))))........)))))))).))..)..))).)))))..))... ( -23.30) >DroSim_CAF1 133025 120 - 1 AAAUAUCAACAGGACACACACGAAAAAAACACCAGGGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUCGGUCAUGUUUGUGUAAUGCGU .........((..(((((.(((........((((((((.((((....((((((((((......))))))))))...))))........))))))))........))).)))))..))... ( -22.49) >DroEre_CAF1 141341 118 - 1 AAAUAUCAACAGGACA--AACGAAAAAAACACCAGAGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUCGGUCAUGUUUGUGUAAUGCGU ..((((.((((.(((.--..((((......((((((((.((((....((((((((((......))))))))))...))))........))))))))))))))).)))).))))....... ( -23.10) >DroYak_CAF1 145176 118 - 1 AAAUAUCAACAGGACA--AACGAAAAGAACACCAGGGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUUGGUCAUGUUUGUGUAAUGCGU ...........(.(((--((((....(((.((((((((.((((....((((((((((......))))))))))...))))........))))))))))).....))))))).)....... ( -20.60) >consensus AAAUAUCAACAGGACA__CACGAAAAAAACACCAGGGAAUAAUGAAAAAAGUUUUAUAAUUGCAUAAAACUUUCCAAUUAACAAGCUGUCUUUGGUUUCGGUCAUGUUUGUGUAAUGCGU ..((((.((((.(((.....((((......((((((((.((((....((((((((((......))))))))))...))))........))))))))))))))).)))).))))....... (-21.22 = -20.82 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:54 2006