
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,253,863 – 17,254,085 |
| Length | 222 |
| Max. P | 0.994889 |

| Location | 17,253,863 – 17,253,965 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -33.46 |
| Consensus MFE | -25.26 |
| Energy contribution | -25.66 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17253863 102 + 23771897 UAUAUAUGUAUGCGGGAGCUGCGGCUGUGGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCA .............((((.(((.(((((.(((..((...(((((((....((((((((...))))))))..)))))))...))..))).))))).)------------------)))))). ( -34.30) >DroSec_CAF1 137661 96 + 1 ------UAUAUGCGGGAGCUGCGGCUGUGGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCA ------.......((((.(((.(((((.(((..((...(((((((....((((((((...))))))))..)))))))...))..))).))))).)------------------)))))). ( -34.30) >DroSim_CAF1 132490 96 + 1 ------UAUAUGCGGGAGCUGCGGCUGUGGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCA ------.......((((.(((.(((((.(((..((...(((((((....((((((((...))))))))..)))))))...))..))).))))).)------------------)))))). ( -34.30) >DroEre_CAF1 140795 90 + 1 ------UAUAUGCGGGAGCUGCGGCUGUUGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCACA------------------------ ------.....((((...))))(((((..((..((...(((((((....((((((((...))))))))..)))))))...))..))..)))))...------------------------ ( -24.40) >DroYak_CAF1 144610 112 + 1 -------AUAUGCG-GAGCUGUGGCUGUUGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCUCAGUCACAGCCACAGCCACAGUCUCA -------.......-(((((((((((((.((........((((((....((((((((...))))))))..))))))..((((((((....))))..))))..)).)))))))))).))). ( -40.00) >consensus ______UAUAUGCGGGAGCUGCGGCUGUGGCACGUAUUUGUUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC__________________AGUCUCA ...........((....))...(((((.(((..((...(((((((....((((((((...))))))))..)))))))...))..))).)))))........................... (-25.26 = -25.66 + 0.40)


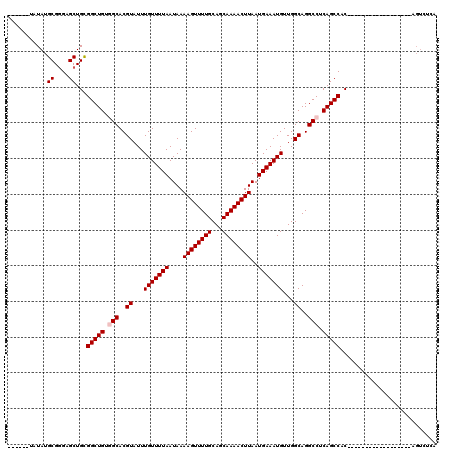
| Location | 17,253,863 – 17,253,965 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.65 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -20.91 |
| Energy contribution | -21.51 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.44 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17253863 102 - 23771897 UGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCCACAGCCGCAGCUCCCGCAUACAUAUAUA .(((.((------------------(((((((.(((.((.............((((((((...))))))))...............)).))).))))))))))))............... ( -30.19) >DroSec_CAF1 137661 96 - 1 UGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCCACAGCCGCAGCUCCCGCAUAUA------ .(((.((------------------(((((((.(((.((.............((((((((...))))))))...............)).))).)))))))))))).........------ ( -30.19) >DroSim_CAF1 132490 96 - 1 UGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCCACAGCCGCAGCUCCCGCAUAUA------ .(((.((------------------(((((((.(((.((.............((((((((...))))))))...............)).))).)))))))))))).........------ ( -30.19) >DroEre_CAF1 140795 90 - 1 ------------------------UGUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCAACAGCCGCAGCUCCCGCAUAUA------ ------------------------((((((((..((.((.............((((((((...))))))))...............)).))..))))))))((....)).....------ ( -21.69) >DroYak_CAF1 144610 112 - 1 UGAGACUGUGGCUGUGGCUGUGACUGAGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCAACAGCCACAGCUC-CGCAUAU------- .(((.((((((((((.((((((..((((((....))))..............((((((((...)))))))).........))..)))).)).)))))))))))))-.......------- ( -37.90) >consensus UGAGACU__________________GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAACAAAUACGUGCCACAGCCGCAGCUCCCGCAUAUA______ .........................(((((((.(((.((.............((((((((...))))))))...............)).))).))))))).((....))........... (-20.91 = -21.51 + 0.60)


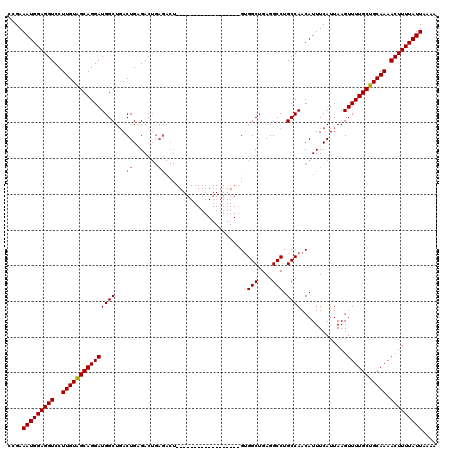
| Location | 17,253,903 – 17,254,005 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17253903 102 + 23771897 UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCAGUCUCAGUCAGCCAUCCUGCUACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((.((((.(((....)))..------------------.(.((.......)).).))))((((......))))....))))))). ( -23.90) >DroSec_CAF1 137695 102 + 1 UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCAGUCGCAGUGAGCCAUCCUGCUACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((.((((........)))).------------------.((((..((.((((.((....)))))).))..))))...))))))). ( -27.60) >DroSim_CAF1 132524 102 + 1 UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC------------------AGUCUCAGUCGCAGUCAGCCAUCCUGCUACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((.((((........)))).------------------.((((..((.((((.........)))).))..))))...))))))). ( -24.80) >DroEre_CAF1 140829 96 + 1 UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCACA------------------------CUCCCAGUCAGCCAUCCUGCCACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((.((((.(((....)))..(------------------------(.....))..))))((((......))))....))))))). ( -23.20) >DroYak_CAF1 144642 120 + 1 UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCUCAGUCACAGCCACAGCCACAGUCUCAGUCUCAGUCAGCCAUCCUGCUACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((.((((((((....))))..((.......)).))))........((((..((.(((......)))))..))))...))))))). ( -25.90) >consensus UUUUAAUAAAAGUUUUGCAGCAAAACUUAAUGAAAUGUUGGCAGGCCUCAGCCAC__________________AGUCUCAGUCGCAGUCAGCCAUCCUGCUACAAGGACCUCCAUUUCGG .........((((((((...))))))))..(((((((..((..(((....))).........................................((((......))))))..))))))). (-21.70 = -21.70 + -0.00)



| Location | 17,253,903 – 17,254,005 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.49 |
| Mean single sequence MFE | -31.78 |
| Consensus MFE | -23.87 |
| Energy contribution | -24.11 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17253903 102 - 23771897 CCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGACUGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..((((((((((((((((.....)).))((((.(------------------(((((........))).))))))).....)))))))))))).))))))))).... ( -29.10) >DroSec_CAF1 137695 102 - 1 CCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUCACUGCGACUGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..((((((((((((.((.....))...(((((.(------------------(((((........))).))))))))....)))))))))))).))))))))).... ( -31.90) >DroSim_CAF1 132524 102 - 1 CCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGCGACUGAGACU------------------GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..((((((((((((.((.....))...(((((.(------------------(((((........))).))))))))....)))))))))))).))))))))).... ( -31.90) >DroEre_CAF1 140829 96 - 1 CCGAAAUGGAGGUCCUUGUGGCAGGAUGGCUGACUGGGAG------------------------UGUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..(((..(((((((...(((...((((------------------------((((((........))).))))))).))).)))))))..))).))))))))).... ( -31.20) >DroYak_CAF1 144642 120 - 1 CCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGACUGAGACUGUGGCUGUGGCUGUGACUGAGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..((((((((((((((((.....)).))((((.((((((...((((..(.(....))...)))).))).))))))).....)))))))))))).))))))))).... ( -34.80) >consensus CCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGACUGAGACU__________________GUGGCUGAGGCCUGCCAACAUUUCAUUAAGUUUUGCUGCAAAACUUUUAUUAAAA ....(((((((((..(((((((((((((((...((.......)).......................(((....))).)))).............))))))))))).))))))))).... (-23.87 = -24.11 + 0.24)

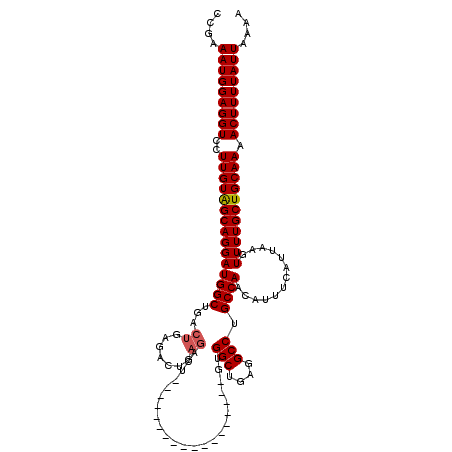
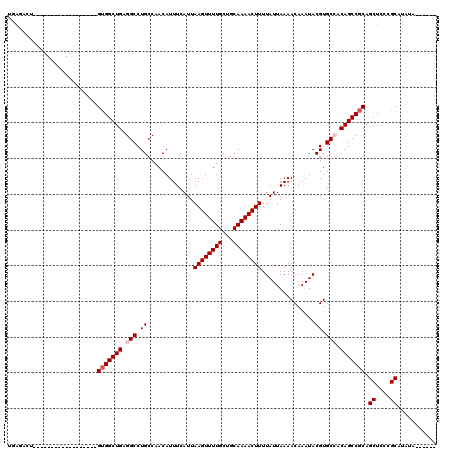
| Location | 17,253,965 – 17,254,085 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -36.30 |
| Energy contribution | -37.70 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17253965 120 - 23771897 CAUUUGCACACACAUAACUUUGCAGCAGUCGCAGUCGGAACAUGUUCAUGUCUUGGGCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGAC ...(((((............)))))..(((.(((((((......(((((...(((((((((.((.(....).)).))))))))).))))).(((((......)))))..))))))).))) ( -42.60) >DroSec_CAF1 137757 120 - 1 CAUUUGCACACACAUAACUUUGCAGCAGUCGCAGUCGGAACAUGUUCAUGUCUUGGGCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUCACUGCGAC ...(((((............)))))..((((((((.((......(((((...(((((((((.((.(....).)).))))))))).))))).(((((......)))))..)).)))))))) ( -42.80) >DroSim_CAF1 132586 120 - 1 CAUUUGCACACACAUAACUUUGCAGCAAUCGCAGUCGGAACAUGUUCCUGUCUUGGGCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGCGAC ...(((((............)))))...((((((((((.......((((((.(((((((((.((.(....).)).)))))))))...((....)).....))))))...)))))))))). ( -47.20) >DroEre_CAF1 140885 120 - 1 CAUUUGCACACACAUAACUUUGCAGCAGUCGCAGUCGGAACGUGUUCAUGUCUUGGUCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUGGCAGGAUGGCUGACUGGGAG ...(((((............)))))...((.(((((((.((((....))))....((((((((..(....)..((.(((.((........)).))).))))))))))..))))))).)). ( -36.80) >DroYak_CAF1 144722 120 - 1 CAUUUGCACACACAUAAGUAAGCAGCAGUCGGAGUCGGAACAUGUUCAUGUCUUGGGCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGAC ...((((..((......))..))))..(((..((((((......(((((...(((((((((.((.(....).)).))))))))).))))).(((((......)))))..))))))..))) ( -40.70) >consensus CAUUUGCACACACAUAACUUUGCAGCAGUCGCAGUCGGAACAUGUUCAUGUCUUGGGCCUGUCUGGGAAACAAGCUAGGCCCGAAAUGGAGGUCCUUGUAGCAGGAUGGCUGACUGAGAC ...(((((............)))))..(((.(((((((......(((((...(((((((((.((.(....).)).))))))))).))))).(((((......)))))..))))))).))) (-36.30 = -37.70 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:51 2006