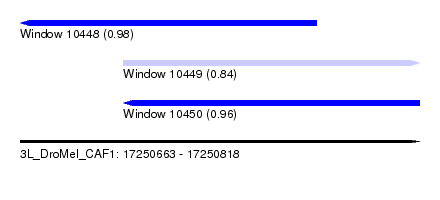
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,250,663 – 17,250,818 |
| Length | 155 |
| Max. P | 0.984351 |

| Location | 17,250,663 – 17,250,778 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.76 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -22.60 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17250663 115 - 23771897 UCGUCAUGGCCCAUGGUACACUGUA-----AUACACCAUUCACGGCCAUGACACGAUACGGUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUAUAGCGAACAGUUUGCCUCAUUUUUC ..(((((((((.(((((........-----....)))))....))))))))).......(((((..........)))))..................((((.....)))).......... ( -25.90) >DroSec_CAF1 134470 115 - 1 UCGUCAUGGCCCAUGGUCCACUGUA-----AUACACCAUUCACGGCCAUGACACGGUACGCUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUUGAGCGAACAGUUUGCCUCAUUUUUC ..(((((((((.(((((........-----....)))))....)))))))))..((((((((((......((((.((....)).))))......)))))).......))))......... ( -30.61) >DroSim_CAF1 129273 115 - 1 UCGUCAUGGCCCAUGGUCCACUGUA-----AUACACCAUUCACGGCCAUGACACGGUACGCUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUUGAGCGAACAGUUUGGCUCAUUUUUC ..(((((((((.(((((........-----....)))))....)))))))))...((.((((((......((((.((....)).))))......)))))).))................. ( -30.20) >DroEre_CAF1 137633 115 - 1 UCGUCAUGGCCCAUGCUCCACUGUA-----CUGCACCAUUCACGCCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUUGAGCGAACAGUUUUCCUCAUUUUUC ..(((((((....(((......)))-----..((.........)))))))))..((.(((((((..........))))).(((..(((.......)))..))).))...))......... ( -19.30) >DroYak_CAF1 141318 120 - 1 UCGUCAUGGCCCAUGGUCCACUGUAUAGUAAUACACCAUUCACGGCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUUGAGCGAGCAGUUUCCCUCAUUUUUC ..(((((((((.(((((.....((((....)))))))))....)))))))))..((.(((((((..........))))).(((..(((.......)))..))).))..)).......... ( -29.50) >consensus UCGUCAUGGCCCAUGGUCCACUGUA_____AUACACCAUUCACGGCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUAGUUUAUUUUCCCUUUGAGCGAACAGUUUGCCUCAUUUUUC ..(((((((((.((((.....((((......))))))))....)))))))))..((((........))))((((((....(((((.........)))))...))))))............ (-22.60 = -23.08 + 0.48)



| Location | 17,250,703 – 17,250,818 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -27.38 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17250703 115 + 23771897 UAGUCAAUUUGAUAUAUGACCGUAUCGUGUCAUGGCCGUGAAUGGUGUAU-----UACAGUGUACCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG .(((((....(((((......)))))(((((((((((....(((((((((-----....))))))))).)))))))))..........(((((.....)))))))........))))).. ( -34.10) >DroSec_CAF1 134510 115 + 1 UAGUCAAUUUGAUAUAUGAGCGUACCGUGUCAUGGCCGUGAAUGGUGUAU-----UACAGUGGACCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG ...(((.((((....((.(((((.....(((((((((....(((((.(((-----....))).))))).)))))))))((..........)).))).)).))..)))).)))........ ( -32.40) >DroSim_CAF1 129313 115 + 1 UAGUCAAUUUGAUAUAUGAGCGUACCGUGUCAUGGCCGUGAAUGGUGUAU-----UACAGUGGACCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG ...(((.((((....((.(((((.....(((((((((....(((((.(((-----....))).))))).)))))))))((..........)).))).)).))..)))).)))........ ( -32.40) >DroEre_CAF1 137673 115 + 1 UAGUCAAUUUGAUAUAUGACCGUACCGUGUCAUGGGCGUGAAUGGUGCAG-----UACAGUGGAGCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG .(((((..........((((.(((((((.((((....))))))))))).)-----).)).....((.(((..(.....)..)))....(((((.....)))))))........))))).. ( -25.10) >DroYak_CAF1 141358 120 + 1 UAGUCAAUUUGAUAUAUGACCGUACCGUGUCAUGGCCGUGAAUGGUGUAUUACUAUACAGUGGACCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG .(((((.....................((((((((((....(((((.((((.......)))).))))).)))))))))).........(((((.....)))))..........))))).. ( -31.90) >consensus UAGUCAAUUUGAUAUAUGACCGUACCGUGUCAUGGCCGUGAAUGGUGUAU_____UACAGUGGACCAUGGGCCAUGACGAACCAAUUAAAUCAACGUCUGAUUGCAAAAUGAAUGACUUG .(((((.....................((((((((((....(((((.(((.........))).))))).)))))))))).........(((((.....)))))..........))))).. (-27.38 = -27.82 + 0.44)

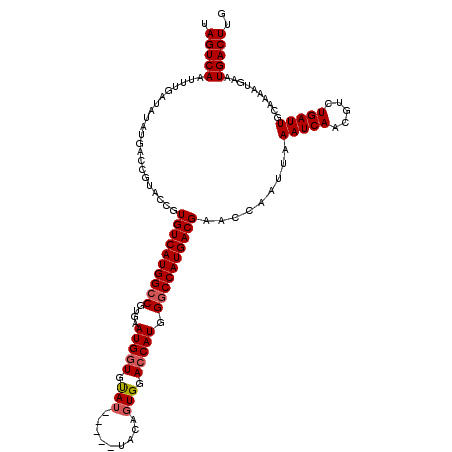
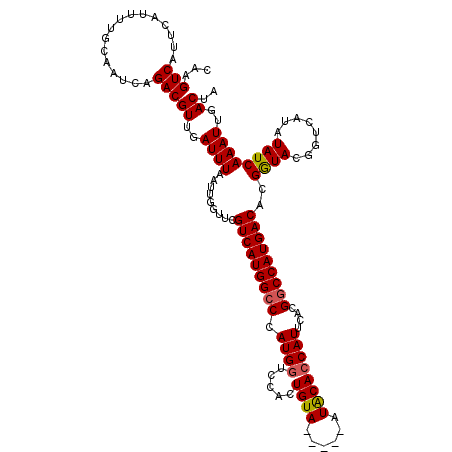
| Location | 17,250,703 – 17,250,818 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -25.19 |
| Energy contribution | -25.27 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17250703 115 - 23771897 CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGGUACACUGUA-----AUACACCAUUCACGGCCAUGACACGAUACGGUCAUAUAUCAAAUUGACUA ...(((................)))((..((((.........(((((((((.(((((........-----....)))))....)))))))))..((((........))))))))..)).. ( -27.89) >DroSec_CAF1 134510 115 - 1 CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGGUCCACUGUA-----AUACACCAUUCACGGCCAUGACACGGUACGCUCAUAUAUCAAAUUGACUA ...(((................)))((..((((.........(((((((((.(((((........-----....)))))....)))))))))..((((........))))))))..)).. ( -27.09) >DroSim_CAF1 129313 115 - 1 CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGGUCCACUGUA-----AUACACCAUUCACGGCCAUGACACGGUACGCUCAUAUAUCAAAUUGACUA ...(((................)))((..((((.........(((((((((.(((((........-----....)))))....)))))))))..((((........))))))))..)).. ( -27.09) >DroEre_CAF1 137673 115 - 1 CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGCUCCACUGUA-----CUGCACCAUUCACGCCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUA ..(((((........(((.(((((((..((((.....)))))))).)))....)))...((((((-----(((...(((........)))...)))))))))............))))). ( -21.50) >DroYak_CAF1 141358 120 - 1 CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGGUCCACUGUAUAGUAAUACACCAUUCACGGCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUA ...(((................)))((..((((.........(((((((((.(((((.....((((....)))))))))....)))))))))..((((........))))))))..)).. ( -28.69) >consensus CAAGUCAUUCAUUUUGCAAUCAGACGUUGAUUUAAUUGGUUCGUCAUGGCCCAUGGUCCACUGUA_____AUACACCAUUCACGGCCAUGACACGGUACGGUCAUAUAUCAAAUUGACUA ...(((................)))((..((((.........(((((((((.((((.....((((......))))))))....)))))))))..((((........))))))))..)).. (-25.19 = -25.27 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:41 2006