| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,920,966 – 1,921,081 |
| Length | 115 |
| Max. P | 0.791458 |

| Location | 1,920,966 – 1,921,081 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -40.98 |
| Consensus MFE | -25.96 |
| Energy contribution | -28.02 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
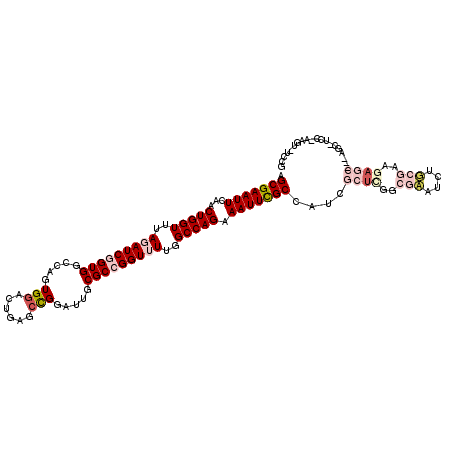
>3L_DroMel_CAF1 1920966 115 + 23771897 AGCGAAUUCAACUGGUUUAGAUCGGUGGCCAUUGGACUGAGCCGGAUUGCGCCGGUUUGGGCCAGAAAUUCGCCAUCGCUCGGCGAAUCUUCGAAGAGC---AGCUUCC-AAGUCUCCG .(((((((...((((((((((((((((....((((......))))....)))))))))))))))).)))))))....((((..(((....)))..))))---.......-......... ( -43.10) >DroPse_CAF1 50566 101 + 1 AGCGAAUUCAACUGGUUUAUAUCAGUGGCUUGUGGCUUGAGCCGGAUUGCGCCGGUUUUGGCCAGAAAUUUGCCAUCCAU---CGCAUCUGAAGAGAGC---UA------------CUG .((((.....((((((....))))))(((((.(((((.(((((((......))))))).))))).))....))).....)---))).(((....)))..---..------------... ( -35.40) >DroEre_CAF1 44895 115 + 1 AGCGAAUUCAACUGGUUUAGAUCGGUGGCCAUUGGACUGAGCCGGAUUGCGCCGGUUUUGGCCAGAAAUUCGCCAUCGCUCGGCGAAUCUUCGAAGAGC---AGCAUCC-AAGUCUCCG .(((((((...(((((..(((((((((....((((......))))....)))))))))..))))).)))))))....((((..(((....)))..))))---.......-......... ( -40.20) >DroYak_CAF1 44377 115 + 1 AGCGAAUUUAACUGGUUUAGAUCGGUGGCCAUUGGACUGAGCCGGAUUGCGCCGGUUUUGGCCAGAAAUUCGCCAUCGCUCGGCGAAUCUUCGAAGAGC---AGCUUCC-AAGUGGCCG .((((((((..(((((..(((((((((....((((......))))....)))))))))..)))))))))))))....((((..(((....)))..))))---.(((...-....))).. ( -42.60) >DroAna_CAF1 47316 119 + 1 AGCGAAUUCAACUGGUUUAGAUCGGUGACCAGUGAACUGAGCUGGAUUGCGCCGGUUUUGGCCAGAAAUUCGCCAUUGCCUGGCGAAGAUGCGGAGAGCGCCAGCCUCCCACUUUUCAG .(((((((...(((((..(((((((((.(((((.......)))))....)))))))))..))))).)))))))......((((.((((.((.((((.((....)))))))))))))))) ( -49.20) >DroPer_CAF1 49818 101 + 1 AGCGAAUUCAACUGGUUUAUAUCAGUGGCUUGUGGCUUGAGCCGGAUUGCGCCGGUUUUGGCCAGAAAUUUGCCAUCCAU---CGCAUCUGAAGAGAGC---UA------------CUG .((((.....((((((....))))))(((((.(((((.(((((((......))))))).))))).))....))).....)---))).(((....)))..---..------------... ( -35.40) >consensus AGCGAAUUCAACUGGUUUAGAUCGGUGGCCAGUGGACUGAGCCGGAUUGCGCCGGUUUUGGCCAGAAAUUCGCCAUCGCUCGGCGAAUCUGCGAAGAGC___AGC_UCC_AAGU_UCCG .(((((((...(((((..(((((((((.....(((......))).....)))))))))..))))).)))))))....((((..(((....)))..)))).................... (-25.96 = -28.02 + 2.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:33 2006