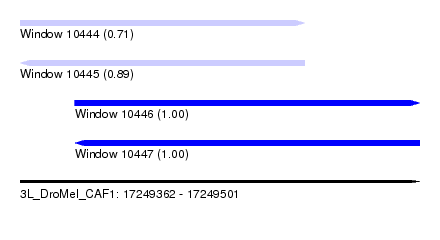
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,249,362 – 17,249,501 |
| Length | 139 |
| Max. P | 0.999906 |

| Location | 17,249,362 – 17,249,461 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -23.02 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17249362 99 + 23771897 UCUUAUCCGGCACCACCC---------------------AACGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU ........(((.......---------------------...))).....(((((...)))))((((((((((..(((((.(((((....)))..)).)))))..))))..))))))... ( -23.50) >DroSec_CAF1 133147 99 + 1 UCUUAUCCGGCACCACCC---------------------AACGCCCGCUUUUGGCCAAGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU .(((((..((......))---------------------...((((((((.(((((((((......)))))......)))).)))))...((....))))).......)))))....... ( -23.60) >DroSim_CAF1 127967 99 + 1 UCUUAUCCGGCACCACCC---------------------AGCGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU ........(((.(((...---------------------(((....)))..)))....)))..((((((((((..(((((.(((((....)))..)).)))))..))))..))))))... ( -26.30) >DroEre_CAF1 136388 106 + 1 UCUUAUCCGCCACCACCCA--------------CCAUACACCACCCACUGUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU ........(((........--------------............((((..(((((((((......)))))......))))..))))...((....)))))................... ( -21.00) >DroYak_CAF1 140014 120 + 1 UCUUAUCCAACACCACCCACAAUCCACAACCCACCACCCACCGCCCACUGUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU ..................................................(((((...)))))((((((((((..(((((.(((((....)))..)).)))))..))))..))))))... ( -20.70) >consensus UCUUAUCCGGCACCACCC_____________________AACGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUU ..................................................(((((...)))))((((((((((..(((((.(((((....)))..)).)))))..))))..))))))... (-20.70 = -20.70 + -0.00)

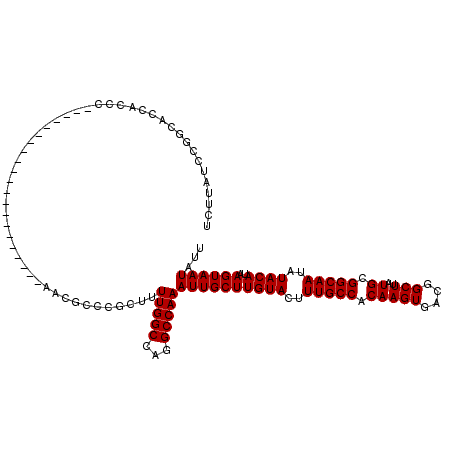
| Location | 17,249,362 – 17,249,461 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.31 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -26.94 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.893070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17249362 99 - 23771897 AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGUU---------------------GGGUGGUGCCGGAUAAGA .......(((((((((.((((((((............)))))))).))))).....((((((.(.(((...(((....)))---------------------.))).).)))))))))). ( -31.40) >DroSec_CAF1 133147 99 - 1 AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCUUGGCCAAAAGCGGGCGUU---------------------GGGUGGUGCCGGAUAAGA .......(((((((((.((((((((............)))))))).))))).....((((((.(.(((...(((....)))---------------------.))).).)))))))))). ( -31.40) >DroSim_CAF1 127967 99 - 1 AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGCU---------------------GGGUGGUGCCGGAUAAGA .......(((((((((.((((((((............)))))))).))))..((..((((((...)))))).)).((((((---------------------....))))))..))))). ( -34.10) >DroEre_CAF1 136388 106 - 1 AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAACAGUGGGUGGUGUAUGG--------------UGGGUGGUGGCGGAUAAGA ..((((((((((((((((..((((....))...((((..((((..((..((((...))))..)).))))..))))))..)))))).)--------------))))))))).......... ( -30.70) >DroYak_CAF1 140014 120 - 1 AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAACAGUGGGCGGUGGGUGGUGGGUUGUGGAUUGUGGGUGGUGUUGGAUAAGA ((((((((((((((((.((((((((............)))))))).))))..(((((..(.(((.(((..(....)..))).)).).)..))))).....))))))))))))........ ( -37.90) >consensus AAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGGU_____________________GGGUGGUGCCGGAUAAGA ..((((((((.(((((.((((((((............)))))))).))))).........(((((........)))))........................)))))))).......... (-26.94 = -26.38 + -0.56)

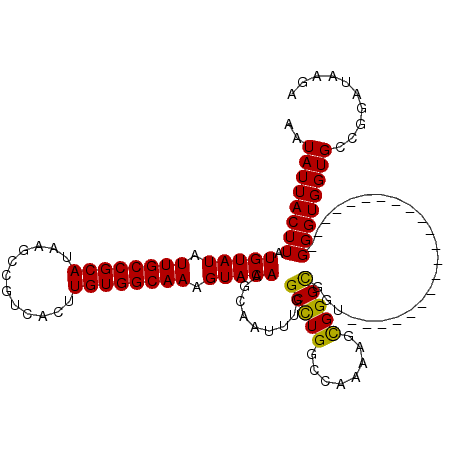
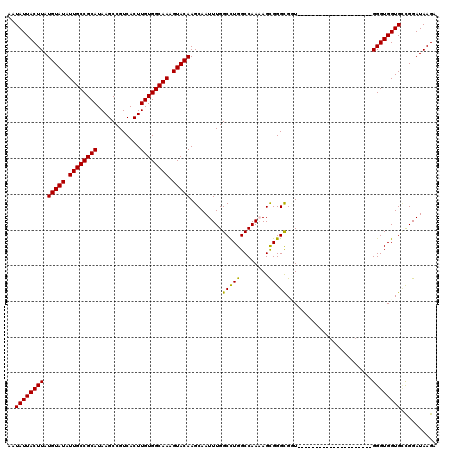
| Location | 17,249,381 – 17,249,501 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -43.78 |
| Energy contribution | -43.78 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17249381 120 + 23771897 ACGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAUGGGCGUUGUCUGGCCUUGGGUGGCGAAUGAG .((((.((((..((((((((....(..(((((........)))))..)..(((((((..(((((((((....)).)))))))....)))))))...))))))))..))))))))...... ( -48.10) >DroSec_CAF1 133166 120 + 1 ACGCCCGCUUUUGGCCAAGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAUGGGCGUUGUCUGGCCUUGGGUGGCGAAUGAG .((((.((((..(((((..(....(..(((((........)))))..)..(((((((..(((((((((....)).)))))))....)))))))...)..)))))..))))))))...... ( -43.20) >DroSim_CAF1 127986 120 + 1 GCGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAUGGGCGUUGUCUGGCCUUGGGUGGCGAAUGAG .((((.((((..((((((((....(..(((((........)))))..)..(((((((..(((((((((....)).)))))))....)))))))...))))))))..))))))))...... ( -47.80) >DroEre_CAF1 136414 120 + 1 CCACCCACUGUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAUGGGCGUUGUCUGGCCUUGGGUGGCGAAAGGG (((((((.....((((((((....(..(((((........)))))..)..(((((((..(((((((((....)).)))))))....)))))))...)))))))).)))))))(....).. ( -48.80) >DroYak_CAF1 140054 120 + 1 CCGCCCACUGUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAGGGGCGUUGUCUGGCCUUGGGUGGCGAACGAG (((((((.....((((((((......((((((........))))))((((.(((.((((.((((((((....)).))))))))))...))).)))))))))))).)))))))........ ( -46.10) >consensus ACGCCCGCUUUUGGCCAGGCCAAAUUGCUUGUACUUUGCCACAAGUGACGGCUUAUGCGGCAAUAUACAUAAGUAAUAUUGCGUACCAUGGGCGUUGUCUGGCCUUGGGUGGCGAAUGAG .((((((.....((((((((....(..(((((........)))))..)..(((((((..(((((((((....)).)))))))....)))))))...)))))))).))))))......... (-43.78 = -43.78 + 0.00)

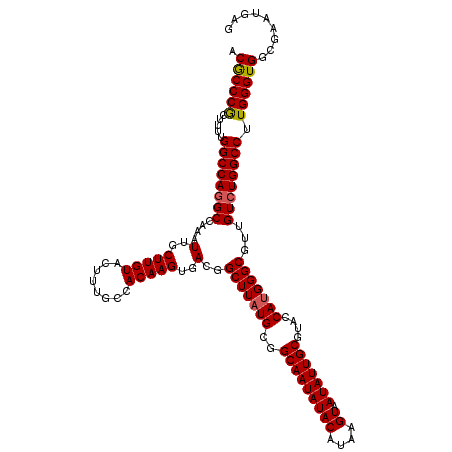
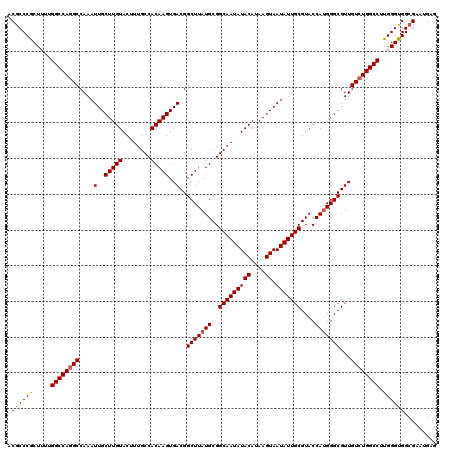
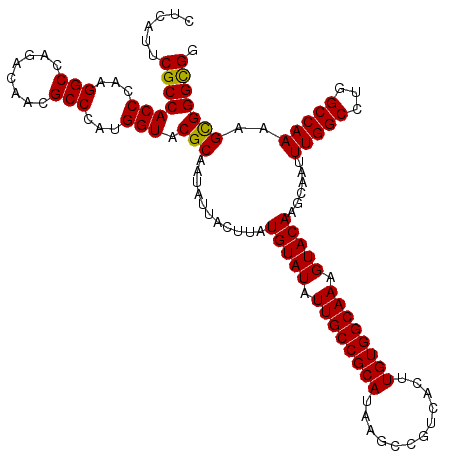
| Location | 17,249,381 – 17,249,501 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.42 |
| Mean single sequence MFE | -40.38 |
| Consensus MFE | -37.68 |
| Energy contribution | -37.28 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.10 |
| SVM RNA-class probability | 0.999798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17249381 120 - 23771897 CUCAUUCGCCACCCAAGGCCAGACAACGCCCAUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGU .......(((......)))......((((((...(((((((((((.((....))))))))).......((((.(....)))))...))))..((..((((((...)))))).)))))))) ( -39.40) >DroSec_CAF1 133166 120 - 1 CUCAUUCGCCACCCAAGGCCAGACAACGCCCAUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCUUGGCCAAAAGCGGGCGU .......(((......)))......((((((...(((((((((((.((....))))))))).......((((.(....)))))...))))..((..((((((...)))))).)))))))) ( -39.40) >DroSim_CAF1 127986 120 - 1 CUCAUUCGCCACCCAAGGCCAGACAACGCCCAUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGC ......(((((((...(((........)))...))).(((...........(((((.((((((((............)))))))).))))).....((((((...)))))).))))))). ( -39.10) >DroEre_CAF1 136414 120 - 1 CCCUUUCGCCACCCAAGGCCAGACAACGCCCAUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAACAGUGGGUGG ........(((((((.((((((.....(((...)))..((...(((.((..(((((.((((((((............)))))))).))))))).)))...)))))))).....))))))) ( -42.70) >DroYak_CAF1 140054 120 - 1 CUCGUUCGCCACCCAAGGCCAGACAACGCCCCUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAACAGUGGGCGG ...(((.(((......)))..)))..(((((((((((((((((((.((....))))))))).......((((.(....)))))...)))).......(((((...)))))))).))))). ( -41.30) >consensus CUCAUUCGCCACCCAAGGCCAGACAACGCCCAUGGUACGCAAUAUUACUUAUGUAUAUUGCCGCAUAAGCCGUCACUUGUGGCAAAGUACAAGCAAUUUGGCCUGGCCAAAAGCGGGCGG ......(((((((...(((........)))...))).(((...........(((((.((((((((............)))))))).)))))......(((((...)))))..))))))). (-37.68 = -37.28 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:38 2006