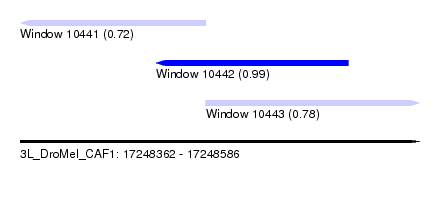
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,248,362 – 17,248,586 |
| Length | 224 |
| Max. P | 0.993727 |

| Location | 17,248,362 – 17,248,466 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.16 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -28.71 |
| Energy contribution | -28.95 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17248362 104 - 23771897 CGCCAUCAAGUGGCAUCC-------UUCGCAUCCU---------GCAGCAUCCUAGGAUAGCGAACUUGGGCCGAGUGGCAGUAAGCUACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU .(((((....((((....-------((((((((((---------(........)))))).))))).....)))).)))))..((((((.((((((.....)))))).))))))....... ( -38.70) >DroSec_CAF1 132148 104 - 1 CGCCAUCAAGUGGCAUCC-------UUCGCAUCCU---------GCAGCAACCGAGGAUGGCGAAGUCGGGCCGAGUUGCAGUAAGCUACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU .(((((...)))))...(-------((((((((((---------..........))))).))))))...(.(((((...((((...(.....).))))..))))).)............. ( -33.80) >DroSim_CAF1 126969 104 - 1 CGCCAUCAAGUGGCAUCC-------UUCGCAUCCU---------GCAGCAACCGAGGAUGGCGAAGUCGGGCCGAGUGGCAGUAAGCUACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU .(((((....((((...(-------((((((((((---------..........))))).))))))....)))).)))))..((((((.((((((.....)))))).))))))....... ( -40.30) >DroEre_CAF1 135392 111 - 1 CGCCAUCAAGUGGCACCCACCGCCAUCCGCAUCCU---------CGAGCUUCCGAGGAUGGCGAAGCCGGCCCGAGUGGCAGUAAGCUACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU ((((.....(((((.......)))))....(((((---------((......)))))))))))((((..(((((((...((((...(.....).))))..))))))).))))........ ( -45.90) >DroYak_CAF1 138979 120 - 1 CGCCAUCAAGUGGCAUCCACCUCCGUCCGCAUCUCCCAGGAAGACGAGCUCCCAAGGAAGGCGAAGUCAGGCCGAGUGGCAGUAAGCCACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU .(((((...)))))..........((((((...(((..((.((.....)).))..)))..))((..((((..((((((((.....))))))))..))))..))))))............. ( -42.40) >consensus CGCCAUCAAGUGGCAUCC_______UUCGCAUCCU_________GCAGCAACCGAGGAUGGCGAAGUCGGGCCGAGUGGCAGUAAGCUACUUGAGCUGAUCUCGGGCAGCUUAAACUUUU .(((((...))))).(((..........(((((((...................))))).))((.(((((.(((((((((.....)))))))).)))))).))))).............. (-28.71 = -28.95 + 0.24)

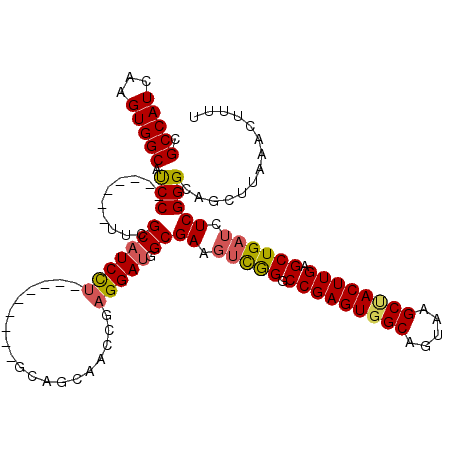
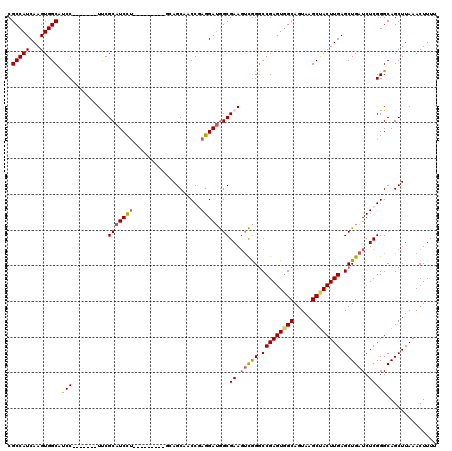
| Location | 17,248,438 – 17,248,546 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.56 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17248438 108 - 23771897 CCUGGAUGCUUGUCAUUGAAACAAAUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAUCCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCAUCC-------UUCGCAUCCU----- ...((((((.................((.(((.....)))..........................((((....).)))))(((((...)))))....-------...)))))).----- ( -22.00) >DroSec_CAF1 132224 108 - 1 CCUGGAUGCUUGUCAUUGAAACAAGUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAGUCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCAUCC-------UUCGCAUCCU----- ...(((((((((.(.((((.....(((((........)))))............)))).).))).(((((....).)))).(((((...)))))....-------...)))))).----- ( -24.63) >DroSim_CAF1 127045 108 - 1 CCUGGAUGCUUGUCAUUGAAACAAAUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAGCCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCAUCC-------UUCGCAUCCU----- ...((((((.................((.(((.....)))..........................((((....).)))))(((((...)))))....-------...)))))).----- ( -22.00) >DroEre_CAF1 135468 115 - 1 CCCGGAUGCAUGUCAUUGAAACAAAUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAGCCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCACCCACCGCCAUCCGCAUCCU----- ...((((((......((((......((((........))))..................((....(((((....).)))).))..))))(((((.......)))))..)))))).----- ( -24.10) >DroYak_CAF1 139059 120 - 1 CCUGGAUGCCUGUCAUUGAAACAAAUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAGCCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCAUCCACCUCCGUCCGCAUCUCCCAGG (((((.(((.((.((((......))))))))).((.((((.........................(((((....).)))).(((((...)))))..............)))).))))))) ( -27.00) >consensus CCUGGAUGCUUGUCAUUGAAACAAAUGCAGCACGAAAUGCACCCAAAACCCCAAUCAAAGCCAAAUUACCUAACGAGUAGCGCCAUCAAGUGGCAUCC_______UUCGCAUCCU_____ ...((((((.................((.(((.....)))..........................((((....).)))))(((((...)))))..............))))))...... (-21.72 = -21.56 + -0.16)

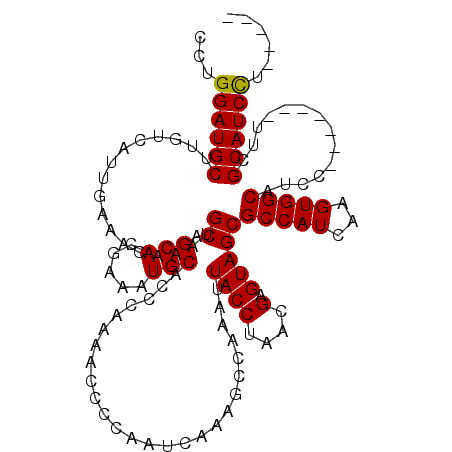

| Location | 17,248,466 – 17,248,586 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.08 |
| Mean single sequence MFE | -34.28 |
| Consensus MFE | -32.94 |
| Energy contribution | -32.86 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.776170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17248466 120 + 23771897 CUACUCGUUAGGUAAUUUGGAUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCAUUUGUUUCAAUGACAAGCAUCCAGGAAAAUCAAAGCUGUGGACUCUGCAAAGUUUCUUGGCUGAU ......((((((.(((((...(((((((....((((((((((...((((...((....))....))))...)))))))))).)))))))..((..(...)..))))))).)))))).... ( -29.50) >DroSec_CAF1 132252 120 + 1 CUACUCGUUAGGUAAUUUGACUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCACUUGUUUCAAUGACAAGCAUCCAGGAAAAUCAAAGUUGUGGACUCUGCAAAGUUCCUUGGCUGAU ......((((((.(((((((((((((((....((((((((((.....(((...)))(((((.....))))))))))))))).))))))))))(..(...)..).))))).)))))).... ( -34.70) >DroSim_CAF1 127073 120 + 1 CUACUCGUUAGGUAAUUUGGCUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCAUUUGUUUCAAUGACAAGCAUCCAGGAAAAUCAAAGUUGUGGACUCUGCAAAGUUCCUUGGCUGAU ......((((((.(((((.(((((((((....((((((((((...((((...((....))....))))...)))))))))).)))))))))((..(...)..))))))).)))))).... ( -33.60) >DroEre_CAF1 135503 120 + 1 CUACUCGUUAGGUAAUUUGGCUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCAUUUGUUUCAAUGACAUGCAUCCGGGAAAAUCAAAGUUGUGGACUCUGCGAAGUUCCUCGGCUGAU ....((((..(((....(((((((((((....((((((((((((.((((...((....))....)))).)))))))))))).)))))))))))...)))..))))(((......)))... ( -37.60) >DroYak_CAF1 139099 120 + 1 CUACUCGUUAGGUAAUUUGGCUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCAUUUGUUUCAAUGACAGGCAUCCAGGAAAAUCAAAGUUGUGGACUCUGCAAAGUUCCUUGGCUGGG ...((.((((((.(((((.(((((((((....((((((((((...((((...((....))....))))...)))))))))).)))))))))((..(...)..))))))).)))))).)). ( -36.00) >consensus CUACUCGUUAGGUAAUUUGGCUUUGAUUGGGGUUUUGGGUGCAUUUCGUGCUGCAUUUGUUUCAAUGACAAGCAUCCAGGAAAAUCAAAGUUGUGGACUCUGCAAAGUUCCUUGGCUGAU ......((((((.(((((((((((((((....((((((((((...((((...((....))....))))...)))))))))).))))))))))(..(...)..).))))).)))))).... (-32.94 = -32.86 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:34 2006