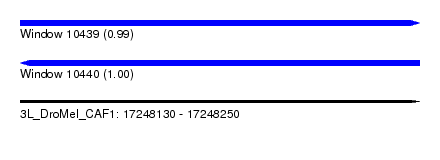
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,248,130 – 17,248,250 |
| Length | 120 |
| Max. P | 0.997277 |

| Location | 17,248,130 – 17,248,250 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -26.77 |
| Energy contribution | -28.48 |
| Covariance contribution | 1.71 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
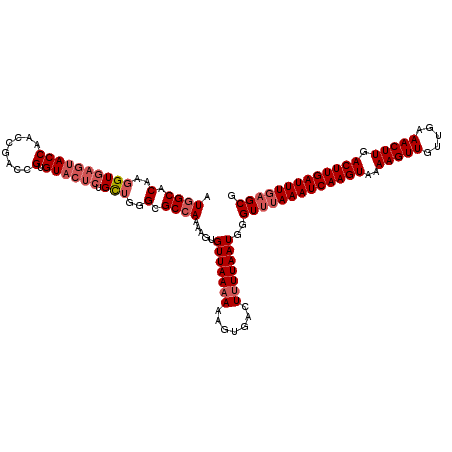
>3L_DroMel_CAF1 17248130 120 + 23771897 AUGGCACAAGGUGAGUACCAACCGACCGUGUACUCUGCUGGGCGCCAAAAGUGUUAAAAAAGUGACUUUUAAUGGGUUUAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUGAGCG .((((.(..((((((((((........).)))))).)))..).)))).....(((((((.......)))))))..(((((((((((((..(((((.....))))).))))))))))))). ( -37.30) >DroSec_CAF1 131916 120 + 1 AUGGCACAAGGUGAGUACCAACCGACCGUGUACUCUGCUGGGCGCCAAAAGUGUUAAAAAAGUGACUUUUAAUGGGUUUAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUCAGCG .((((.(..((((((((((........).)))))).)))..).)))).....(((((((.......)))))))..(((.(((((((((..(((((.....))))).))))))))).))). ( -33.30) >DroSim_CAF1 126745 116 + 1 AUGGCACAA-GUGAGUACCAACCGACCGUGUACUCUGCUGGGCGC-AAAAGUGUUAA-AAAGUGACUUUUAAUGGGU-UAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUGAGCG ..(((((.(-(((((((((........).)))))).))).(((((-....)))))..-...))).)).......(.(-((((((((((..(((((.....))))).))))))))))).). ( -31.10) >DroEre_CAF1 135191 101 + 1 AUGGCACAAGGUGAGU-------------------UGUUGGGCGCCAAAAGUGUUAAAAAAGUGACUUUUAAUGGGUUUAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUGAGCG ......((....((((-------------------..((.(((((.....))))).....))..))))....)).(((((((((((((..(((((.....))))).))))))))))))). ( -27.10) >DroYak_CAF1 138739 120 + 1 AUGGCACAAGGUGAGUACCAACCGACCGUGUACUCUGUUGGGCGCCAAAAGUGUUAAAAAAGUGACUUUUAAUGGGUUUAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUGAGCG .((((((..((((....(((((.((........)).))))).))))....))))))...................(((((((((((((..(((((.....))))).))))))))))))). ( -35.50) >consensus AUGGCACAAGGUGAGUACCAACCGACCGUGUACUCUGCUGGGCGCCAAAAGUGUUAAAAAAGUGACUUUUAAUGGGUUUAAAUCAAGUAAAAGUUGUUGAAACUUGACUUGAUUUGAGCG .((((.(..((((((((((........).)))))).)))..).)))).....(((((((.......)))))))..(((((((((((((..(((((.....))))).))))))))))))). (-26.77 = -28.48 + 1.71)

| Location | 17,248,130 – 17,248,250 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -26.01 |
| Consensus MFE | -21.39 |
| Energy contribution | -22.84 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
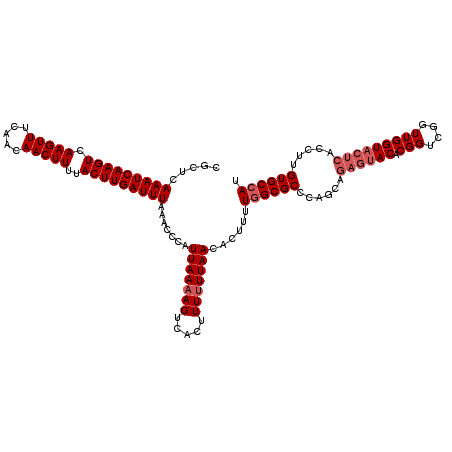
>3L_DroMel_CAF1 17248130 120 - 23771897 CGCUCAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUAAACCCAUUAAAAGUCACUUUUUUAACACUUUUGGCGCCCAGCAGAGUACACGGUCGGUUGGUACUCACCUUGUGCCAU .....(((((((((.(((((.....)))))..))))))))).......(((((((.....)))))))......((((((..((..((((((.(((....)))))))))..)).)))))). ( -28.10) >DroSec_CAF1 131916 120 - 1 CGCUGAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUAAACCCAUUAAAAGUCACUUUUUUAACACUUUUGGCGCCCAGCAGAGUACACGGUCGGUUGGUACUCACCUUGUGCCAU .(((((((((((((.(((((.....)))))..))))))))).......((((((((............))))))))....)))).(.(((((.(((.(((....))).))).)))))).. ( -30.10) >DroSim_CAF1 126745 116 - 1 CGCUCAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUA-ACCCAUUAAAAGUCACUUU-UUAACACUUUU-GCGCCCAGCAGAGUACACGGUCGGUUGGUACUCAC-UUGUGCCAU .....(((((((((.(((((.....)))))..)))))))))(-(((..(((((((....)))-)))).(((((.-((.....)))))))........))))(((((....-..))))).. ( -26.10) >DroEre_CAF1 135191 101 - 1 CGCUCAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUAAACCCAUUAAAAGUCACUUUUUUAACACUUUUGGCGCCCAACA-------------------ACUCACCUUGUGCCAU .....(((((((((.(((((.....)))))..))))))))).......(((((((.....)))))))......((((((......-------------------.........)))))). ( -18.16) >DroYak_CAF1 138739 120 - 1 CGCUCAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUAAACCCAUUAAAAGUCACUUUUUUAACACUUUUGGCGCCCAACAGAGUACACGGUCGGUUGGUACUCACCUUGUGCCAU .....(((((((((.(((((.....)))))..))))))))).......(((((((.....)))))))......((((((......((((((.(((....))))))))).....)))))). ( -27.60) >consensus CGCUCAAAUCAAGUCAAGUUUCAACAACUUUUACUUGAUUUAAACCCAUUAAAAGUCACUUUUUUAACACUUUUGGCGCCCAGCAGAGUACACGGUCGGUUGGUACUCACCUUGUGCCAU .....(((((((((.(((((.....)))))..))))))))).......(((((((.....)))))))......((((((......((((((.(((....))))))))).....)))))). (-21.39 = -22.84 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:31 2006