
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,246,165 – 17,246,495 |
| Length | 330 |
| Max. P | 0.962339 |

| Location | 17,246,165 – 17,246,263 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.11 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246165 98 + 23771897 -CCGCUCUCCCGCUAUCCCACAUCCCACCUCUCGUCCUUG-----UCGUUUGCCGUUUGCCACCUUAAUUGGCUGUGGCCAGGCAAACGCAGCACGAUGGAGCA -..(((((...(........)..........((((.((.(-----.((((((((....(((((((.....))..)))))..))))))))))).)))).))))). ( -29.60) >DroSec_CAF1 130021 90 + 1 -CCGCUCUC--------CCACAUCCCACCUCUCGUCCUUG-----UCGUUUGCCGUUUGCCACCUUAAUUGGCUGUGGCCAGGCAAACGCAGCACGAUGGGGCA -........--------................(.(((..-----((((((((.(((((((........((((....))))))))))))))).))))..)))). ( -31.30) >DroSim_CAF1 123891 90 + 1 -CCGCUCUC--------CCACAUCCCACCUCUCGUUCUUG-----UCGUUUGCCGUUUGCCACCUUAAUUGGCUGUGGCCAGGCAAACGCAGCACGAUGGGGCA -........--------..........((((((((.((.(-----.((((((((....(((((((.....))..)))))..))))))))))).)))).)))).. ( -30.80) >DroEre_CAF1 133206 96 + 1 UCCCUCCUC--------CCGCCUCCCACCUCUCGUCCUUGCCGCUUCGUUUGCCGUUUGCCACCUUAAUUGGCUGUGGCCAGACAAACGCAGCAGGAUGGAGCA .........--------...........(((.((((((.((.((((.((((((((...((((.......))))..))).))))).)).)).))))))))))).. ( -29.40) >consensus _CCGCUCUC________CCACAUCCCACCUCUCGUCCUUG_____UCGUUUGCCGUUUGCCACCUUAAUUGGCUGUGGCCAGGCAAACGCAGCACGAUGGAGCA ............................(((.((((..........((((((((....(((((((.....))..)))))..))))))))......))))))).. (-21.30 = -21.11 + -0.19)



| Location | 17,246,165 – 17,246,263 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 87.97 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.62 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.723038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246165 98 - 23771897 UGCUCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA-----CAAGGACGAGAGGUGGGAUGUGGGAUAGCGGGAGAGCGG- ..((((.((((.((.((((((((..(((((...........)))))....)))))))).-----..)).)))).........(((......))))))).....- ( -34.70) >DroSec_CAF1 130021 90 - 1 UGCCCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA-----CAAGGACGAGAGGUGGGAUGUGG--------GAGAGCGG- ..(((((((.(((((((((((((..(((((...........)))))....)))))))).-----))....(....)))).)))).))--------).......- ( -34.40) >DroSim_CAF1 123891 90 - 1 UGCCCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA-----CAAGAACGAGAGGUGGGAUGUGG--------GAGAGCGG- ..(((((((.(((((((((((((..(((((...........)))))....)))))))).-----))....(....)))).)))).))--------).......- ( -34.40) >DroEre_CAF1 133206 96 - 1 UGCUCCAUCCUGCUGCGUUUGUCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGAAGCGGCAAGGACGAGAGGUGGGAGGCGG--------GAGGAGGGA ..((((.(((((((((.(((((((.(((.(.((((.......))))...((.....))..).)))..)))))))..))....)))))--------))))))... ( -36.40) >consensus UGCCCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA_____CAAGGACGAGAGGUGGGAUGUGG________GAGAGCGG_ ....(((.(((.(..((((((((..(((((...........)))))....))))))).............(....))..).))))))................. (-27.94 = -27.62 + -0.31)



| Location | 17,246,204 – 17,246,299 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.32 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.14 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246204 95 - 23771897 CACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCUCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA----- (((.......((.....))........(((((((..--------------------.))))))).)))...((((((((..(((((...........)))))....)))))))).----- ( -29.30) >DroSec_CAF1 130052 95 - 1 CACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCCCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA----- (((.......((.....))........(((((((..--------------------.))))))).)))...((((((((..(((((...........)))))....)))))))).----- ( -32.30) >DroSim_CAF1 123922 95 - 1 CACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCCCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA----- (((.......((.....))........(((((((..--------------------.))))))).)))...((((((((..(((((...........)))))....)))))))).----- ( -32.30) >DroEre_CAF1 133238 96 - 1 CACAAUUUGUUGACAUUCAUUAUUUUUGUGGG------------------------UGCUCCAUCCUGCUGCGUUUGUCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGAAGCGG .......((..(.(((((((.......)))))------------------------)))..))..(((((.((((((((..(((((...........)))))....)))))))).))))) ( -30.40) >DroYak_CAF1 136850 116 - 1 CACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUCCGCUCCUUCCUACUCCAUCCCACUCCAUCCUGCUGCGUUUGUCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGAA---- .....(((((((...............((((((....................)))))).......(((..(.((((..((((....))))..)))))..)))..)))))))....---- ( -25.85) >consensus CACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC____________________UGCUCCAUCGUGCUGCGUUUGCCUGGCCACAGCCAAUUAAGGUGGCAAACGGCAAACGA_____ ...........................(((((((.......................))))))).......((((((((..(((((...........)))))....))))))))...... (-26.02 = -26.14 + 0.12)


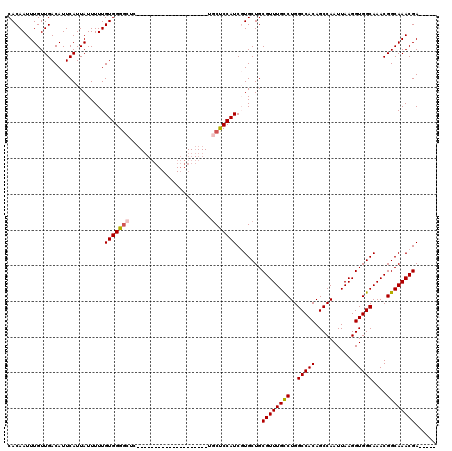
| Location | 17,246,239 – 17,246,339 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.70 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.56 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246239 100 - 23771897 CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCUCCAUCGUGCUGCGUUUGCCU ..(.((..((((((......(((((((..(((.....(((((........))))).)))..)))))))((((((..--------------------.))))))..)))..)))..)).). ( -20.60) >DroSec_CAF1 130087 100 - 1 CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCCCCAUCGUGCUGCGUUUGCCU ..(.((..((((((......(((((((..(((.....(((((........))))).)))..)))))))((((((..--------------------.))))))..)))..)))..)).). ( -23.60) >DroSim_CAF1 123957 100 - 1 CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC--------------------UGCCCCAUCGUGCUGCGUUUGCCU ..(.((..((((((......(((((((..(((.....(((((........))))).)))..)))))))((((((..--------------------.))))))..)))..)))..)).). ( -23.60) >DroEre_CAF1 133278 96 - 1 CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGG------------------------UGCUCCAUCCUGCUGCGUUUGUCU ..(.((..(((.........(((((((..(((.....(((((........))))).)))..))))))).(((------------------------((...)))))....)))..)).). ( -18.10) >DroYak_CAF1 136886 120 - 1 CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUCCGCUCCUUCCUACUCCAUCCCACUCCAUCCUGCUGCGUUUGUCU ..(.((..(((..(((....(((((((..(((.....(((((........))))).)))..))))))).(((((...))))).......................)))..)))..)).). ( -21.50) >consensus CUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC____________________UGCUCCAUCGUGCUGCGUUUGCCU ..(.((..(((.........(((((((..(((.....(((((........))))).)))..)))))))((((((.......................)))))).......)))..)).). (-19.20 = -19.56 + 0.36)



| Location | 17,246,263 – 17,246,379 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.20 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -21.12 |
| Energy contribution | -21.32 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246263 116 - 23771897 GCGUCUUUUUCAUUUUUCGGUUUAUGCGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC---- ..................(((....(((.((....)).)))........)))..(((((((((((((..(((.....(((((........))))).)))..))))))).)))))).---- ( -23.10) >DroSec_CAF1 130111 116 - 1 GCGUCCUUUUAAUUUUUCGGUUUAUGCGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC---- ..(((((...........(((....(((.((....)).)))........)))........(((((((..(((.....(((((........))))).)))..))))))).)))))..---- ( -23.70) >DroSim_CAF1 123981 116 - 1 GCGUCCUUUUCAUUUUUCGGUUUAUGCGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC---- ..(((((...........(((....(((.((....)).)))........)))........(((((((..(((.....(((((........))))).)))..))))))).)))))..---- ( -23.70) >DroEre_CAF1 133302 107 - 1 -----GCCAUCAUUUUUUGGCUUAUGUGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGG-------- -----((((........))))(((((.((.....(((....))).....))))))).((.(((((((..(((.....(((((........))))).)))..))))))).)).-------- ( -22.60) >DroYak_CAF1 136926 115 - 1 -----CUUUUCAUUUUUCGGUUUGUGUGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUCCGCU -----.............(((..(((.((((....))..))...)))..)))..(((((((((((((..(((.....(((((........))))).)))..))))))).))))))..... ( -25.20) >consensus GCGUCCUUUUCAUUUUUCGGUUUAUGCGCUGUCCACAUCGCUGUCACUUGCCAUAGUCUUCAGAAAUUUUGAUUUCUUGUCACAAUUUGUUGACAUUCAUUAUUUUUGUGGGGCUC____ ..................(((....(((.((....)).)))........)))..(((((((((((((..(((.....(((((........))))).)))..))))))).))))))..... (-21.12 = -21.32 + 0.20)

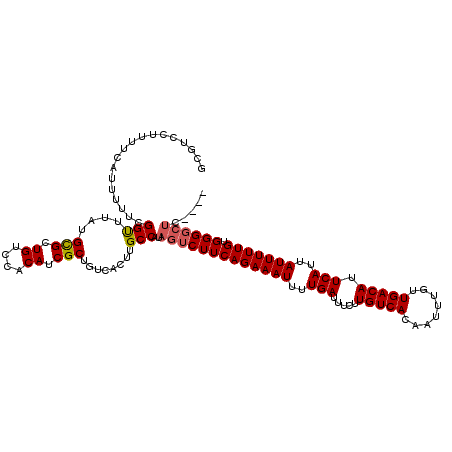

| Location | 17,246,379 – 17,246,495 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -27.20 |
| Consensus MFE | -20.02 |
| Energy contribution | -19.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246379 116 + 23771897 AGGACACAGGAAGCGGAUGCGAAAGGUGC--GCAGGGAAAAUACCCUGUGUGGGAAAAUAUUUUAAUUAAAAUGGAAUAUUAAAAUGUAUGCCAAUAUAUUGCCAACUAUAUUUGUAA ............(((((((.....(((((--((((((......)))))))).((...((((((((((...........))))))))))...))........))).....))))))).. ( -27.10) >DroSec_CAF1 130227 113 + 1 AGGACACAGGAAGUGGAUGCGAAAGGUGCUCACAGGGAAAAUACCCUGUGCGGGAAAUAAUUUUAAUUAAAAUGGAAUAUUUAAAUGUAUUCCAAUGCAGU-----AUAUAUGUGUAA ...(((((....(((..((((.......(.(((((((......))))))).)....................((((((((......)))))))).))))..-----.))).))))).. ( -30.20) >DroSim_CAF1 124097 111 + 1 AGGACACAGGAAGUGAAUGGGAAAGGUGA--GCAGGGAAAAUACCCUGUGCGGGAAAUAAUUUUAAUUAAAAUGGAAUAUUUAAAUGUAUUCCAAUGCAGU-----AUAUAUGUGUAA ...(((((.....................--((((((......))))))(((.....((((....))))...((((((((......)))))))).)))...-----.....))))).. ( -24.30) >consensus AGGACACAGGAAGUGGAUGCGAAAGGUGC__GCAGGGAAAAUACCCUGUGCGGGAAAUAAUUUUAAUUAAAAUGGAAUAUUUAAAUGUAUUCCAAUGCAGU_____AUAUAUGUGUAA ...(((((.......................((((((......))))))(((.......(((((....)))))(((((((......)))))))..))).............))))).. (-20.02 = -19.80 + -0.22)



| Location | 17,246,379 – 17,246,495 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -14.57 |
| Energy contribution | -15.23 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.67 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17246379 116 - 23771897 UUACAAAUAUAGUUGGCAAUAUAUUGGCAUACAUUUUAAUAUUCCAUUUUAAUUAAAAUAUUUUCCCACACAGGGUAUUUUCCCUGC--GCACCUUUCGCAUCCGCUUCCUGUGUCCU ......((((((..(((.......(((.....((((((((...........))))))))......)))..(((((......))))).--((.......))....)))..))))))... ( -20.90) >DroSec_CAF1 130227 113 - 1 UUACACAUAUAU-----ACUGCAUUGGAAUACAUUUAAAUAUUCCAUUUUAAUUAAAAUUAUUUCCCGCACAGGGUAUUUUCCCUGUGAGCACCUUUCGCAUCCACUUCCUGUGUCCU ..(((((.....-----..(((..(((((((........)))))))......................(((((((......)))))))..........))).........)))))... ( -24.29) >DroSim_CAF1 124097 111 - 1 UUACACAUAUAU-----ACUGCAUUGGAAUACAUUUAAAUAUUCCAUUUUAAUUAAAAUUAUUUCCCGCACAGGGUAUUUUCCCUGC--UCACCUUUCCCAUUCACUUCCUGUGUCCU ..(((((.....-----..(((..(((((((........)))))))...((((....))))......)))(((((......))))).--.....................)))))... ( -19.00) >consensus UUACACAUAUAU_____ACUGCAUUGGAAUACAUUUAAAUAUUCCAUUUUAAUUAAAAUUAUUUCCCGCACAGGGUAUUUUCCCUGC__GCACCUUUCGCAUCCACUUCCUGUGUCCU ..(((((.................(((((((........)))))))........................(((((......)))))........................)))))... (-14.57 = -15.23 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:28 2006