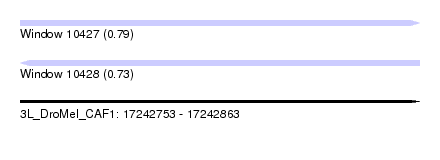
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,242,753 – 17,242,863 |
| Length | 110 |
| Max. P | 0.789171 |

| Location | 17,242,753 – 17,242,863 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -25.09 |
| Energy contribution | -25.69 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242753 110 + 23771897 GGCCAUAAAAAAG-----UGAGCAGGCAGGACAAGAAAGGCUUGAAACAAAAAGGCCGGCUAUGC-----UGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCGAA .............-----(((((((((.(((((((.....))))..((((((((((((((...))-----)).))))))).))))))((((....))))....)))......)))))).. ( -33.40) >DroSec_CAF1 126617 110 + 1 GGCCAUAAAAAAG-----UGAGCAGGCAGGACAAGAAAGGCUUGAAACAAAAAGGAAGGCUGUGC-----UGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCGCA ............(-----(((((((((.(((((((.....))))..(((....(((((((.....-----...))))))).))))))((((....))))....)))......))))))). ( -33.40) >DroSim_CAF1 120512 110 + 1 GGCCAUAAAAAAG-----UGAGCAGGCAGGACAAGAAAGGCUUGAAACAAAAAGGCCGGCUAUGC-----UGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCGAA .............-----(((((((((.(((((((.....))))..((((((((((((((...))-----)).))))))).))))))((((....))))....)))......)))))).. ( -33.40) >DroEre_CAF1 129759 107 + 1 GGCCAUAAAAAAG-----UGAGCAGGC-GGAC--GAAAGGCUUGAAACAAAAAGGCCGGCUAUUC-----CGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCCAA ............(-----.((((((((-((((--....).......(((((((((((((.....)-----)).))))))).))))))((((....))))....)))......)))))).. ( -31.40) >DroYak_CAF1 133299 118 + 1 GGCCAUAAAAAAGCAAGUUGAGCUGGCAGGAC--GAAAGGCUUGAAACAAAAAGGCCGCCUAUGCUGUGCUGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCGAA .................((((((((((.((((--(((((((.....(((.(.(((...))).)..))).....))))))))...)))((((....))))....))))......)))))). ( -31.60) >consensus GGCCAUAAAAAAG_____UGAGCAGGCAGGACAAGAAAGGCUUGAAACAAAAAGGCCGGCUAUGC_____UGUGCCUUUUGUGUUCCGACACGACUGUCAACAGCCAUUAAAUGCUCGAA ...................((((((((.(((((.(((((((............((....))............))))))).))))).((((....))))....)))......)))))... (-25.09 = -25.69 + 0.60)

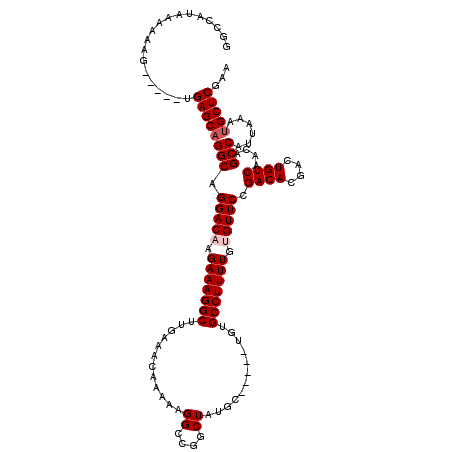
| Location | 17,242,753 – 17,242,863 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -26.55 |
| Energy contribution | -26.75 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242753 110 - 23771897 UUCGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACA-----GCAUAGCCGGCCUUUUUGUUUCAAGCCUUUCUUGUCCUGCCUGCUCA-----CUUUUUUAUGGCC ...(((((....(((((((....)))))))...((((((.(((((((.(.-----((...)).))))))))))))))...................))))).-----............. ( -30.80) >DroSec_CAF1 126617 110 - 1 UGCGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACA-----GCACAGCCUUCCUUUUUGUUUCAAGCCUUUCUUGUCCUGCCUGCUCA-----CUUUUUUAUGGCC ...(((((....(((((((....)))))))((.(((.((..((((((...-----..((((.........)))).....))))))..))))).)).))))).-----............. ( -27.90) >DroSim_CAF1 120512 110 - 1 UUCGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACA-----GCAUAGCCGGCCUUUUUGUUUCAAGCCUUUCUUGUCCUGCCUGCUCA-----CUUUUUUAUGGCC ...(((((....(((((((....)))))))...((((((.(((((((.(.-----((...)).))))))))))))))...................))))).-----............. ( -30.80) >DroEre_CAF1 129759 107 - 1 UUGGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACG-----GAAUAGCCGGCCUUUUUGUUUCAAGCCUUUC--GUCC-GCCUGCUCA-----CUUUUUUAUGGCC ...(((((....(((((((....)))))))...((((((.(((((((.((-----(.....)))))))))))))))).........--....-...))))).-----............. ( -34.60) >DroYak_CAF1 133299 118 - 1 UUCGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACAGCACAGCAUAGGCGGCCUUUUUGUUUCAAGCCUUUC--GUCCUGCCAGCUCAACUUGCUUUUUUAUGGCC ...((((.....(((((((....)))))))...((((((.(((((((...((.(......))).))))))))))))).........--.........)))).....(((.......))). ( -33.30) >consensus UUCGAGCAUUUAAUGGCUGUUGACAGUCGUGUCGGAACACAAAAGGCACA_____GCAUAGCCGGCCUUUUUGUUUCAAGCCUUUCUUGUCCUGCCUGCUCA_____CUUUUUUAUGGCC ...((((.....(((((((....)))))))...((((((.(((((((.................)))))))))))))....................))))................... (-26.55 = -26.75 + 0.20)

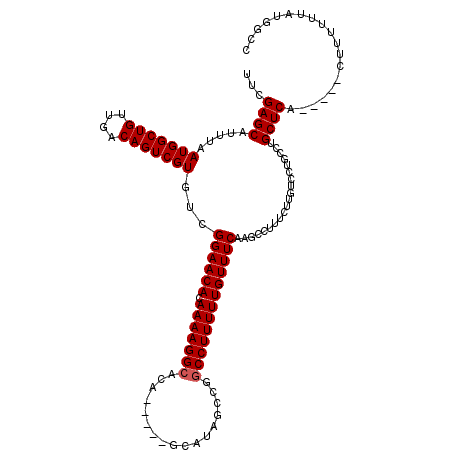
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:20 2006