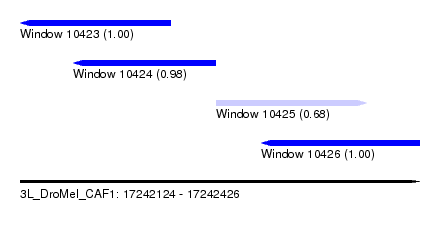
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,242,124 – 17,242,426 |
| Length | 302 |
| Max. P | 0.998467 |

| Location | 17,242,124 – 17,242,238 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.02 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -24.08 |
| Energy contribution | -24.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242124 114 - 23771897 ------CCCAUUUCCAUUACCAUCACAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUGCGAUUUGAACUUUCACUUGAU ------.................((((((((.(((.(((.............))).))).)))))))).(((..(((((((((((((.......))).)))).))).)))..)))..... ( -27.12) >DroSec_CAF1 125994 114 - 1 ------CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUGCGAUUUGAACUUUCACUUGAU ------................((((((((((..................(((((.....)))))))))))))))(((((.((..((..(((((((....))))))).))..)).))))) ( -25.86) >DroSim_CAF1 119889 114 - 1 ------CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUGCGAUUUGAACUUUCACUUGAU ------................((((((((((..................(((((.....)))))))))))))))(((((.((..((..(((((((....))))))).))..)).))))) ( -25.86) >DroEre_CAF1 129115 116 - 1 ----CAGCCAUUUCCAUUUCCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGCUGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUCCGAUUUGAACUUUCACUUGAU ----..((((..((.((((((((.(((((((.(((.(((.............))).))).)))))).).)))))))).))....)))).(((((((....)))))))............. ( -30.32) >DroYak_CAF1 132643 120 - 1 UCCGCAGAAAUCUCCAUUUCCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUAGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUCCGAUUUGAACUUUCACUUGAU ......(((((....)))))..((((((((((.((((((...((((......))))..)))).))))))))))))(((((.((..((..(((((((....))))))).))..)).))))) ( -30.10) >consensus ______CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGAUUGCUGGCAUUAAAUCAUGCGAUUUGAACUUUCACUUGAU ......................((((((((((........((((((......)).))))......))))))))))(((((.((..((..(((((((....))))))).))..)).))))) (-24.08 = -24.28 + 0.20)


| Location | 17,242,164 – 17,242,272 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.51 |
| Mean single sequence MFE | -21.32 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984188 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242164 108 - 23771897 AACCCGACAUACAACACCCAU------UUUCAAUCCCAAA------CCCAUUUCCAUUACCAUCACAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGA ....((((............(------((........)))------.............((((((((((((.(((.(((.............))).))).)))))))).))))..)))). ( -20.82) >DroSec_CAF1 126034 108 - 1 AACCCGACAUACAACACCCAU------UUUCAAUCCCAAA------CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGA ....((((............(------((........)))------................((((((((((..................(((((.....))))))))))))))))))). ( -19.86) >DroSim_CAF1 119929 108 - 1 AACCCGACAUACAACACCCAU------UUUCAAUCCCAAA------CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGA ....((((............(------((........)))------................((((((((((..................(((((.....))))))))))))))))))). ( -19.86) >DroEre_CAF1 129155 113 - 1 AACCCGACGUCCAACACCCUUUGCCAUUGCCAAUCCC-------CAGCCAUUUCCAUUUCCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGCUGCAUGUGCGUGGGAGUCGA ....((((..............((....)).......-------..................((((((((((..................(((((.....))))))))))))))))))). ( -22.86) >DroYak_CAF1 132683 116 - 1 ----CAACACCCUUUACCAUUCGCAAUCCCCAAUCCCCAAUCCGCAGAAAUCUCCAUUUCCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUAGCAUGUGCGUGGGAGUCGA ----................((((......................(((((....)))))..((((((((((.((((((...((((......))))..)))).))))))))))))).))) ( -23.20) >consensus AACCCGACAUACAACACCCAU______UUUCAAUCCCAAA______CCCAUUUCCAUUACCAUCCCAUGCACCAUCUAUUCAUUAUUAUUAUGUAAAUGUUGCAUGUGCGUGGGAGUCGA ..............................................................((((((((((........((((((......)).))))......))))))))))..... (-15.78 = -15.98 + 0.20)

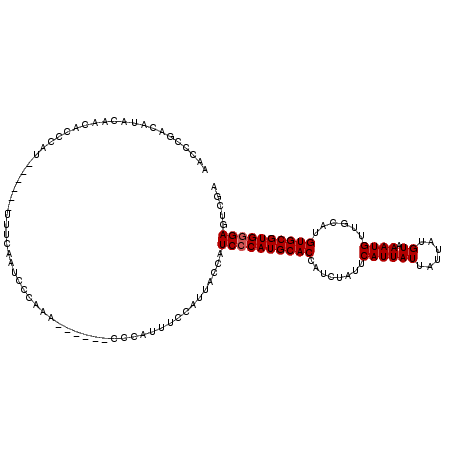
| Location | 17,242,272 – 17,242,386 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.77 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -27.78 |
| Energy contribution | -27.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242272 114 + 23771897 UUGGUAGUGGUAG------UGGUAGUGGUAGUGGGCCUGGCCAGCAUAAUUCAUUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU .....(((((...------.(((((.(((.(..(((((((((.((((((..........)))))))))))((((((((.........)))))).))))))..)))).)))))...))))) ( -31.60) >DroSec_CAF1 126142 108 + 1 UUGGCAGUGGU------------AGUGGUAGUGGGCCUGGCCAGCAUAAUUCAAUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU ..(((((....------------(((((....((((((((((.((((((..........)))))))))))((((((((.........)))))).)))))))))))).)))))........ ( -29.30) >DroSim_CAF1 120037 108 + 1 UUGGCAGUGGU------------AGUGGUAGUGGGCCUGGCCAGCAUAAUUCAAUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU ..(((((....------------(((((....((((((((((.((((((..........)))))))))))((((((((.........)))))).)))))))))))).)))))........ ( -29.30) >DroEre_CAF1 129268 120 + 1 UUGGUAGUGGUAGUGGCAGUGGCAGUGGCAGUGGGCUUGGCCAGCAUAAUUCAAUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU .....(((((....(((((.(((...((((...((((.((((.((((((..........))))))))))))))..)))).........((.....))......))).)))))...))))) ( -33.70) >DroYak_CAF1 132799 102 + 1 U------------------UGGUAGUGGUAGUGGGCCUGGCCAGCAUAAUUCAAUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU .------------------....(((((....((((((((((.((((((..........)))))))))))).((((((.........)))))).(((.....)))....))))..))))) ( -28.20) >consensus UUGGCAGUGGU________UGG_AGUGGUAGUGGGCCUGGCCAGCAUAAUUCAAUUAAUUUAUGCGGUCGGUCAAUGUUUAGUUUUUACGUUGUGCGGUUUUCGCUUUUGCCUUUUCGCU .......................(((((....((((((((((.((((((..........)))))))))))).((((((.........)))))).(((.....)))....))))..))))) (-27.78 = -27.62 + -0.16)



| Location | 17,242,306 – 17,242,426 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.67 |
| Mean single sequence MFE | -25.90 |
| Consensus MFE | -25.90 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17242306 120 - 23771897 GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAAUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... ( -25.90) >DroSec_CAF1 126170 120 - 1 GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAUUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... ( -25.90) >DroSim_CAF1 120065 120 - 1 GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAUUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... ( -25.90) >DroEre_CAF1 129308 120 - 1 GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAUUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... ( -25.90) >DroYak_CAF1 132821 120 - 1 GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAUUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... ( -25.90) >consensus GGCUCUUGGUCUACGCAUAAAUUAUGCAAAAUGAUUUAUAAGCGAAAAGGCAAAAGCGAAAACCGCACAACGUAAAAACUAAACAUUGACCGACCGCAUAAAUUAAUUGAAUUAUGCUGG .......((((..((((((((((((.....)))))))))..)))....(((((..(((.....))).....((....))......))).))))))((((((.((....)).))))))... (-25.90 = -25.90 + 0.00)

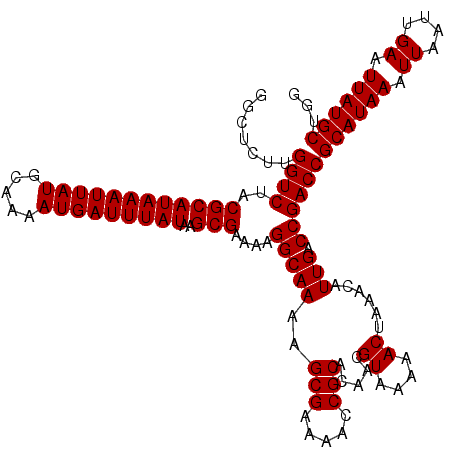

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:18 2006