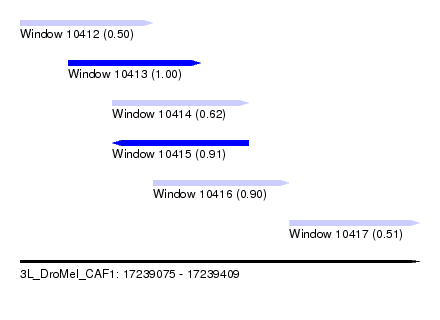
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,239,075 – 17,239,409 |
| Length | 334 |
| Max. P | 0.995647 |

| Location | 17,239,075 – 17,239,186 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -21.61 |
| Energy contribution | -21.89 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239075 111 + 23771897 AAUAAAUUAUGCCUAGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGC .(((((((((((..((((((..((((......((......)).......(((((((....))))))))---))).)))))))))------).)))))))..................... ( -29.70) >DroSec_CAF1 123026 111 + 1 AAUAAAUUAUGCCUAGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGC .(((((((((((..((((((..((((......((......)).......(((((((....))))))))---))).)))))))))------).)))))))..................... ( -29.70) >DroSim_CAF1 116934 111 + 1 AAUAAAUUAUGCCUAGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGC .(((((((((((..((((((..((((......((......)).......(((((((....))))))))---))).)))))))))------).)))))))..................... ( -29.70) >DroEre_CAF1 126229 117 + 1 AAUAAAUUAUGCCUGGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCAGCGCACUCGCGCCGAG---CACUAUUGUUGCAAACUCGUAUAUUUAUGGCACAUUAUGUGUUUUUUGC ....(((..(((.(((((.((((((......))))))...((((((....)))))).....))))).)---))..)))...(((((..((((....))))((((.....))))..))))) ( -30.80) >DroYak_CAF1 129559 114 + 1 AAUAAAUUAUGCCUGGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCAGUGCACCCGCACCGAGUUGCACUAUAGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGC .........((((....(((((((((((((.(((.(((....)))(((.((.((((....)))).))))))))..))))..)))------))))))...))))................. ( -23.90) >consensus AAUAAAUUAUGCCUAGCGAUUUGUGUUUAUUUGCAAACAAGCGUUGACAUCGGUGCACCCGCGCCGAG___CACUAUCGUUGCA______UAAAUUUAUGGCACAUUAUAUGUUUUUUGC .........((((....((((((........(((((.............(((((((....)))))))............)))))......))))))...))))................. (-21.61 = -21.89 + 0.28)


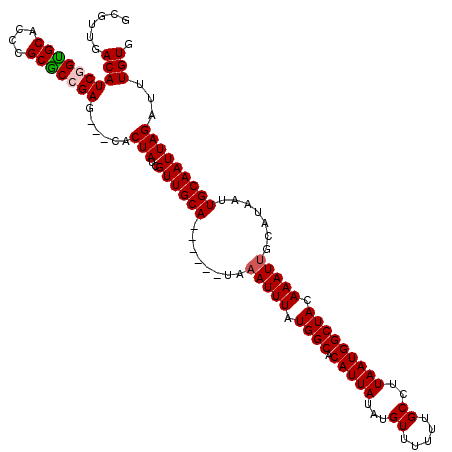
| Location | 17,239,115 – 17,239,226 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.46 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239115 111 + 23771897 GCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG (((......(((((((....))))))).---......)))..((------(((((((.((((.(((((...((.....))..)))))))))..(((((((....)))))))))))))))) ( -32.52) >DroSec_CAF1 123066 111 + 1 GCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG (((......(((((((....))))))).---......)))..((------(((((((.((((.(((((...((.....))..)))))))))..(((((((....)))))))))))))))) ( -32.52) >DroSim_CAF1 116974 111 + 1 GCGUUGACAUCGGUGCACCCGCGCCGAG---CACUAUCGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG (((......(((((((....))))))).---......)))..((------(((((((.((((.(((((...((.....))..)))))))))..(((((((....)))))))))))))))) ( -32.52) >DroEre_CAF1 126269 117 + 1 GCGUUGACAUCAGCGCACUCGCGCCGAG---CACUAUUGUUGCAAACUCGUAUAUUUAUGGCACAUUAUGUGUUUUUUGCCCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG ((((((....))))))...(((((((((---((....))))(((((..((((....))))((((.....))))..))))).....))))....(((((((....)))))))......))) ( -30.50) >DroYak_CAF1 129599 114 + 1 GCGUUGACAUCAGUGCACCCGCACCGAGUUGCACUAUAGUUGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG ((...(((.((.((((....)))).)))))))..........((------(((((((.((((.(((((...((.....))..)))))))))..(((((((....)))))))))))))))) ( -27.00) >consensus GCGUUGACAUCGGUGCACCCGCGCCGAG___CACUAUCGUUGCA______UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUG ......((((((((((....)))))))......(((..((((((........(((((.((((.(((((...((.....))..))))))))).))))).......)))))))))...))). (-26.72 = -27.00 + 0.28)


| Location | 17,239,152 – 17,239,266 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -21.52 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.616544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239152 114 + 23771897 UGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAA ((((------(.......((((((((...)))).....(((((((.((((((((((((((....)))).....)))))).)))).((.....))...))))))).)))).)))))..... ( -27.90) >DroSec_CAF1 123103 114 + 1 UGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAA ((((------(.......((((((((...)))).....(((((((.((((((((((((((....)))).....)))))).)))).((.....))...))))))).)))).)))))..... ( -27.90) >DroSim_CAF1 117011 114 + 1 UGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAA ((((------(.......((((((((...)))).....(((((((.((((((((((((((....)))).....)))))).)))).((.....))...))))))).)))).)))))..... ( -27.90) >DroEre_CAF1 126306 120 + 1 UGCAAACUCGUAUAUUUAUGGCACAUUAUGUGUUUUUUGCCCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGGAAAAUGCAAUUAA .(((..((((((.(((((..(((.(((((((..(((..(((.....)))...)))..))))))))))...))))).))))))..))).....(((((((........)))))))...... ( -29.30) >DroYak_CAF1 129639 114 + 1 UGCA------UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGGCAGCUUAACGCGUUUUGAGGUCGCAAAAUGCAAUUAA ..((------(((((((.((((.(((((...((.....))..)))))))))..(((((((....))))))))))))))))............((((((((......))))))))...... ( -28.20) >consensus UGCA______UAAAUUUAUGGCACAUUAUAUGUUUUUUGCCUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAA ((((.........((((.((((.(((((...((.....))..))))))))).))))))))((((((((.((.(((((..((((..((.....))))))..)))))...)).)))))))). (-21.52 = -21.72 + 0.20)



| Location | 17,239,152 – 17,239,266 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Mean single sequence MFE | -25.54 |
| Consensus MFE | -19.79 |
| Energy contribution | -20.35 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239152 114 - 23771897 UUAAUUGCAUUUGGCGACCUCAAAACGCGUUAAGCAGGCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAAGGCAAAAAACAUAUAAUGUGCCAUAAAUUUA------UGCA .....(((((.((((...........((.((((...((((.((((((.....((((....)))))))))))))).)))).)).....((((...)))))))).......)------)))) ( -26.70) >DroSec_CAF1 123103 114 - 1 UUAAUUGCAUUUGGCGACCUCAAAACGCGUUAAGCAGGCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAAGGCAAAAAACAUAUAAUGUGCCAUAAAUUUA------UGCA .....(((((.((((...........((.((((...((((.((((((.....((((....)))))))))))))).)))).)).....((((...)))))))).......)------)))) ( -26.70) >DroSim_CAF1 117011 114 - 1 UUAAUUGCAUUUGGCGACCUCAAAACGCGUUAAGCAGGCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAAGGCAAAAAACAUAUAAUGUGCCAUAAAUUUA------UGCA .....(((((.((((...........((.((((...((((.((((((.....((((....)))))))))))))).)))).)).....((((...)))))))).......)------)))) ( -26.70) >DroEre_CAF1 126306 120 - 1 UUAAUUGCAUUUUCCGACCUCAAAACGCGUUAAGCAGGCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAGGGCAAAAAACACAUAAUGUGCCAUAAAUAUACGAGUUUGCA (((((.((.((((........)))).)))))))((((((((........(((((((....))))))).((((((.....)))......(((.....))).........))))))))))). ( -26.80) >DroYak_CAF1 129639 114 - 1 UUAAUUGCAUUUUGCGACCUCAAAACGCGUUAAGCUGCCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAGGGCAAAAAACAUAUAAUGUGCCAUAAAUUUA------UGCA .(((((((((((((......))))).((.....)).................))))))))((((((((((.(((((((.(........)...))))).)).))))))...------)))) ( -20.80) >consensus UUAAUUGCAUUUGGCGACCUCAAAACGCGUUAAGCAGGCUCACAAAUCUAAUUGCAAUUAUGCAAUUUGUAGCCAUUAAGGCAAAAAACAUAUAAUGUGCCAUAAAUUUA______UGCA .....((((..((((...........((.((((...((((.((((((.....((((....)))))))))))))).)))).)).....(((.....)))))))..............)))) (-19.79 = -20.35 + 0.56)

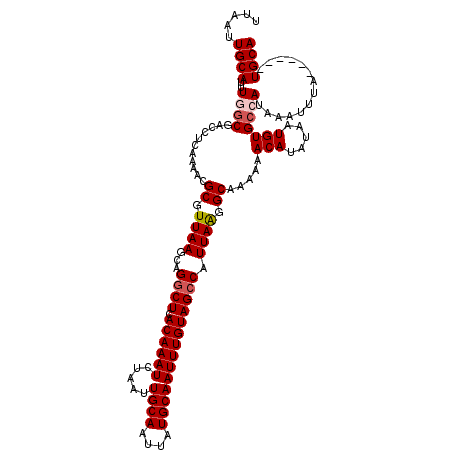
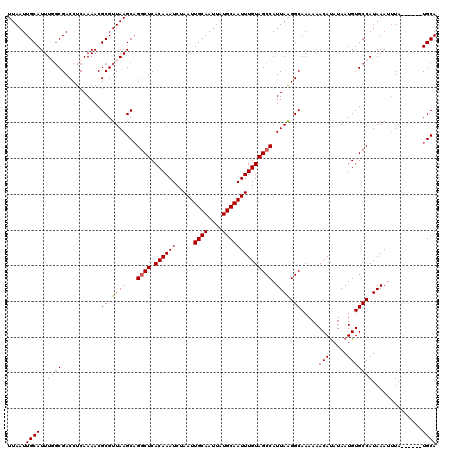
| Location | 17,239,186 – 17,239,300 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239186 114 + 23771897 CUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAAGUAAAACAAUUCG------GUGCGAUGCCAAAUGCCCAAC ......(((....(((((((....))))))).((.((((.....(((((((.((((((.(......).))))))..)))))))..)))).))(------((.....)))....))).... ( -25.80) >DroSec_CAF1 123137 114 + 1 CUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAAGUAACACAAUUCG------GUGCGAUGCCAAAUGCCCAAC ......(((....(((((((....))))))).((.(((((....(((((((.((((((.(......).))))))..))))))).))))).))(------((.....)))....))).... ( -27.90) >DroSim_CAF1 117045 114 + 1 CUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAAGUAACACAAUUCG------GUGCGAUGCCAAAUGCCCAAC ......(((....(((((((....))))))).((.(((((....(((((((.((((((.(......).))))))..))))))).))))).))(------((.....)))....))).... ( -27.90) >DroEre_CAF1 126346 112 + 1 CCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGGAAAAUGCAAUUAAGUAACACAAUUCG------CUGCGAUGCCAAAU-CCC-AC .....((((....(((((((....))))))).((.(((((....(((((((.(((((((........)))))))..))))))).))))).)).------.......))))...-...-.. ( -28.50) >DroYak_CAF1 129673 119 + 1 CCUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGGCAGCUUAACGCGUUUUGAGGUCGCAAAAUGCAAUUAAGUAACACAGUUCGGUGCCGUUGCGAUGCCAAAU-CCCAAC ......(((.((.(((((((....))))))).((.(((((.....((((((.((((((((......))))))))..))))))..))))).)).)))))((((.(((.....))-).)))) ( -33.50) >consensus CUUAAUGGCUACAAAUUGCAUAAUUGCAAUUAGAUUUGUGAGCCUGCUUAACGCGUUUUGAGGUCGCCAAAUGCAAUUAAGUAACACAAUUCG______GUGCGAUGCCAAAUGCCCAAC .....((((....(((((((....))))))).((.(((((....(((((((.((((((.(......).))))))..))))))).))))).))..............)))).......... (-25.48 = -25.92 + 0.44)

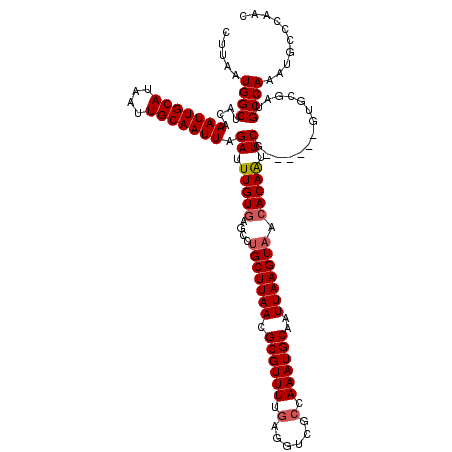
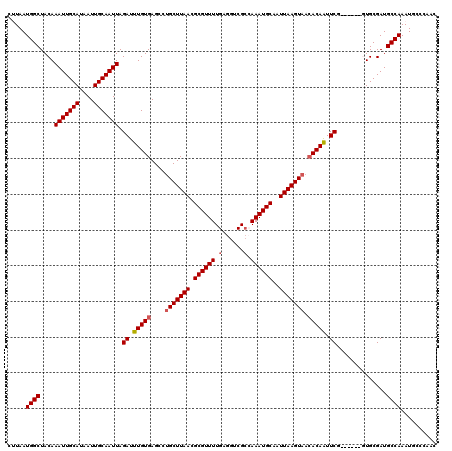
| Location | 17,239,300 – 17,239,409 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.91 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -22.95 |
| Energy contribution | -23.62 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17239300 109 + 23771897 UAGAUCUAUGGCGGGGAGACAA----CGCAUAACCGCUUGGCGG-AAUUUUCCACCAGAAGCCGAAACAAAGGAAAAUGAACGGAAAAUGGUUGCCGAAAAGAAAAGCGGAAAG ..........(((.(....)..----)))....(((((((((((-.(((((((..((....((........))....))...)))))))..)))))........)))))).... ( -29.20) >DroSec_CAF1 123251 109 + 1 UAGAUCUAUGGCGG-UAGACAACGAACGCAUAACCGGUUGGCGGUUAUUUUCCACCAAAAGCCGAAACAAAGGA----GAACGGAAAAUGGGUGCCGAAAAGAAAGGCGGAAAG ....(((((....)-)))).......(((........((((((.(((((((((........((........)).----....))))))))).))))))........)))..... ( -27.21) >DroSim_CAF1 117159 109 + 1 UAGAUCUAUGGCGG-UAGACAACGAACGCAUAACCGGUUGGCGGUUAUUUUCCACCAAAGGCCGAAACAAAGGA----GAACGGAAAAUGGGUGCCGAAAAGAAAGGCGGAAAG ....(((((....)-)))).......(((........((((((.(((((((((.((...))((........)).----....))))))))).))))))........)))..... ( -27.59) >consensus UAGAUCUAUGGCGG_UAGACAACGAACGCAUAACCGGUUGGCGGUUAUUUUCCACCAAAAGCCGAAACAAAGGA____GAACGGAAAAUGGGUGCCGAAAAGAAAGGCGGAAAG ..........................(((........((((((.(((((((((........((........)).........))))))))).))))))........)))..... (-22.95 = -23.62 + 0.67)

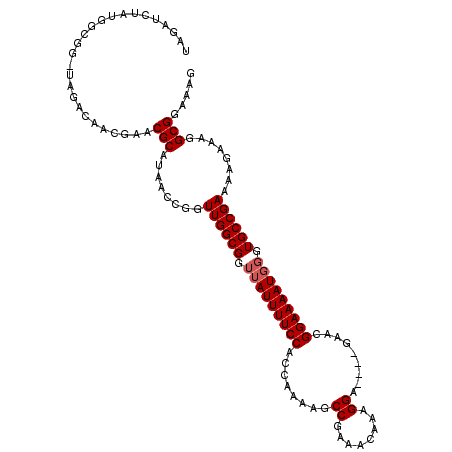
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:09 2006