
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,238,699 – 17,238,937 |
| Length | 238 |
| Max. P | 0.979639 |

| Location | 17,238,699 – 17,238,818 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.57 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.78 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
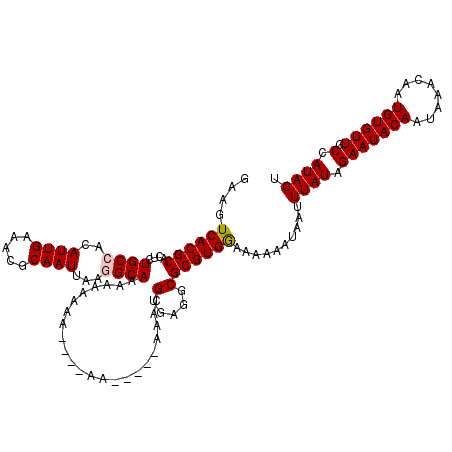
>3L_DroMel_CAF1 17238699 119 + 23771897 AAAGUGUAGCAUACUUAUGAGGGCGAAUA-AAAAAGGUGGCGAAAAUAACCACGCUUUGUUGCACGCGUAAUUUGCAUUAUUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACC .(((((..(((...(((((...((((...-..((((((((.........)))).)))).))))...)))))..)))..)))))((.((((((((((........)))))))))).))... ( -27.70) >DroSec_CAF1 122663 119 + 1 AAAGUGUAGCAUACUUAUGAGGGCGAAUG-AAAAAGGUGGCGAAAAUAACCACGCUUUGUUGCACGCCUAAUUUGCAUUAUUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACC ..((((((((((....))).(((((....-..((((((((.........)))).))))......)))))...)))))))...(((.((((((((((........)))))))))).))).. ( -28.30) >DroSim_CAF1 116581 119 + 1 AAAGUGUGGCAUACUUAUGAGGGCGAAUG-AAAAAGGUGGCGAAAAUAACCACGCUUUGUUGCACGCGUAAUUUGCAUUAUUUGCUAGUAAUUGAAUAAAGUUUUUGAAUUAUUCGCACC ...(((((.............((((((((-(.....((((.........))))((...(((((....)))))..)).)))))))))(((((((.((........)).)))))))))))). ( -27.40) >DroEre_CAF1 125891 120 + 1 AAAGUGCAGCAUACUUAUGAGGGCGAAUGCAAAAAGGUGGCGAAAAUAACCACGCUUUGUUGCACGCGUAAUUUGCAUUAUUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACC ...((((((((.....(((...(((..(((((((((((((.........)))).)))).)))))..)))......)))....))))((((((((((........)))))))))).)))). ( -32.50) >DroYak_CAF1 129175 120 + 1 AAAGUGCAGCAUACUUAUGAGGGCGAGUGAAAAAAGGUGGCGAAAAUAAGCACGCUUUGUUGCACGCGUAAUUUGCAUUAUUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCUCC ...(((((((.(((((........)))))....((((((((........)).)))))))))))))(((.....))).......((.((((((((((........)))))))))).))... ( -32.00) >consensus AAAGUGUAGCAUACUUAUGAGGGCGAAUG_AAAAAGGUGGCGAAAAUAACCACGCUUUGUUGCACGCGUAAUUUGCAUUAUUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACC ...(((((((...((.....))..........((((((((.........)))).)))))))))))(((.....))).......((.((((((((((........)))))))))).))... (-24.58 = -24.78 + 0.20)


| Location | 17,238,699 – 17,238,818 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.57 |
| Mean single sequence MFE | -23.29 |
| Consensus MFE | -19.14 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17238699 119 - 23771897 GGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAAUAAUGCAAAUUACGCGUGCAACAAAGCGUGGUUAUUUUCGCCACCUUUUU-UAUUCGCCCUCAUAAGUAUGCUACACUUU ((.((((((((........................(((..((((....))))....)))...((((.(((((.......))))))))).)-))))))))).................... ( -22.90) >DroSec_CAF1 122663 119 - 1 GGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAAUAAUGCAAAUUAGGCGUGCAACAAAGCGUGGUUAUUUUCGCCACCUUUUU-CAUUCGCCCUCAUAAGUAUGCUACACUUU ((.((((((..........................(((..((((....))))....)))...((((.(((((.......)))))))))..-.)))))))).................... ( -22.00) >DroSim_CAF1 116581 119 - 1 GGUGCGAAUAAUUCAAAAACUUUAUUCAAUUACUAGCAAAUAAUGCAAAUUACGCGUGCAACAAAGCGUGGUUAUUUUCGCCACCUUUUU-CAUUCGCCCUCAUAAGUAUGCCACACUUU (((..((((((..........))))))...((((.(((.....))).......(((......((((.(((((.......)))))))))..-....))).......)))).)))....... ( -22.30) >DroEre_CAF1 125891 120 - 1 GGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAAUAAUGCAAAUUACGCGUGCAACAAAGCGUGGUUAUUUUCGCCACCUUUUUGCAUUCGCCCUCAUAAGUAUGCUGCACUUU (((((((((((..........)))))....((((.(((.....)))......((.((((((.((((.(((((.......))))))))))))))).))........))))...)))))).. ( -26.80) >DroYak_CAF1 129175 120 - 1 GGAGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAAUAAUGCAAAUUACGCGUGCAACAAAGCGUGCUUAUUUUCGCCACCUUUUUUCACUCGCCCUCAUAAGUAUGCUGCACUUU ...((((((((..........))))).........(((.....)))......)))(((((.....((((((((((....((...............))....)))))))))))))))... ( -22.46) >consensus GGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAAUAAUGCAAAUUACGCGUGCAACAAAGCGUGGUUAUUUUCGCCACCUUUUU_CAUUCGCCCUCAUAAGUAUGCUACACUUU ((.((((((..........................(((..((((....))))....)))...((((.(((((.......)))))))))....)))))))).................... (-19.14 = -19.54 + 0.40)


| Location | 17,238,778 – 17,238,897 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17238778 119 + 23771897 UUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACCGAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAAUGUAAA-AAAAAAACGUGAGGCGCGUGGAAAA ..(((.((((((((((........)))))))))).)))......((((((.(((((((..((((.....))))...)))).......((((...-......)))))))..)))))).... ( -27.70) >DroSec_CAF1 122742 120 + 1 UUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACCGAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAACAUCAAUAAAAAAACGUGAUGCGCGUGGAAAA ..(((.((((((((((........)))))))))).)))......((((((....((((..((((.....))))...))))........(((((...........))))).)))))).... ( -27.30) >DroSim_CAF1 116660 116 + 1 UUUGCUAGUAAUUGAAUAAAGUUUUUGAAUUAUUCGCACCGAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAAUAAAAA----UAAAUGUGAGGCGCGUGGAAAA ..(((.(((((((.((........)).))))))).)))......((((((.(((((((..((((.....))))...)))).......(((....----....))))))..)))))).... ( -23.80) >DroEre_CAF1 125971 105 + 1 UUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACCGAAGUCACGCACUCUGCCACAUUGAAAUGCAAUUAAUGCAAUAA---------------AAACGUGGUGCGCGUGGAAAA ..(((.((((((((((........)))))))))).)))......((((((.(...(((((.((....((((.....))))....---------------.)).)))))).)))))).... ( -27.30) >DroYak_CAF1 129255 105 + 1 UUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCUCCGAAGUCACGCACUCUGCCACAUUGAAAUGCAAUUAAUGCAAUAA---------------AAACGUGAGGCGCGUGAAAAA ...((.((((((((((........)))))))))).)).......((((((.....(((.(((.....((((.....))))....---------------....))).))))))))).... ( -23.82) >consensus UUUGCUAGUAAUUUAAUAAAGUUUUUGAAUUAUUCGCACCGAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAA____AA_____AAACGUGAGGCGCGUGGAAAA ...((.((((((((((........)))))))))).)).......((((((....((((..((((.....))))...)))).......................(.....))))))).... (-20.44 = -20.88 + 0.44)


| Location | 17,238,778 – 17,238,897 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.90 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -18.01 |
| Energy contribution | -17.65 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17238778 119 - 23771897 UUUUCCACGCGCCUCACGUUUUUUU-UUUACAUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUCGGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAA .......((((((((((((......-...........(((((((...((((.....))))..))))))).))))))....)))))).................................. ( -25.07) >DroSec_CAF1 122742 120 - 1 UUUUCCACGCGCAUCACGUUUUUUUAUUGAUGUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUCGGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAA .....(((((((...(((((........))))).....((((((...((((.....))))..)))))))))))))((....))((...(((((.((........)).)))))...))... ( -26.70) >DroSim_CAF1 116660 116 - 1 UUUUCCACGCGCCUCACAUUUA----UUUUUAUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUCGGUGCGAAUAAUUCAAAAACUUUAUUCAAUUACUAGCAAA .....(((((((........((----....))......((((((...((((.....))))..)))))))))))))((....))..((((((..........))))))............. ( -23.10) >DroEre_CAF1 125971 105 - 1 UUUUCCACGCGCACCACGUUU---------------UUAUUGCAUUAAUUGCAUUUCAAUGUGGCAGAGUGCGUGACUUCGGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAA .....(((((((.(((((((.---------------....((((.....))))....)))))))....)))))))((....))((...(((((.((........)).)))))...))... ( -26.50) >DroYak_CAF1 129255 105 - 1 UUUUUCACGCGCCUCACGUUU---------------UUAUUGCAUUAAUUGCAUUUCAAUGUGGCAGAGUGCGUGACUUCGGAGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAA ....((((((((((((((((.---------------....((((.....))))....))))))..)).))))))))(....).((...(((((.((........)).)))))...))... ( -25.00) >consensus UUUUCCACGCGCCUCACGUUU_____UU____UUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUCGGUGCGAAUAAUUCAAAAACUUUAUUAAAUUACUAGCAAA .....(((((((.(((((((....................(((.......)))....)))))))....)))))))((....))((...(((((.((........)).)))))...))... (-18.01 = -17.65 + -0.36)



| Location | 17,238,818 – 17,238,937 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -21.62 |
| Consensus MFE | -15.40 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17238818 119 + 23771897 GAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAAUGUAAA-AAAAAAACGUGAGGCGCGUGGAAAAAAUAAUUUAUUGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((.(((((((..((((.....))))...)))).......((((...-......)))))))..)))))).........(((((((....)))))))...((((....)))).. ( -21.70) >DroSec_CAF1 122782 120 + 1 GAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAACAUCAAUAAAAAAACGUGAUGCGCGUGGAAAAAAUAAUUUAUAGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((....((((..((((.....))))...))))........(((((...........))))).))))))..........((((.(((((((........)))))).).)))). ( -20.90) >DroSim_CAF1 116700 116 + 1 GAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAAUAAAAA----UAAAUGUGAGGCGCGUGGAAAAAAUAAUUUAUAGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((.(((((((..((((.....))))...)))).......(((....----....))))))..))))))..........((((.(((((((........)))))).).)))). ( -19.30) >DroEre_CAF1 126011 105 + 1 GAAGUCACGCACUCUGCCACAUUGAAAUGCAAUUAAUGCAAUAA---------------AAACGUGGUGCGCGUGGAAAAAAUAUUUUAUUGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((.(...(((((.((....((((.....))))....---------------.)).)))))).)))))).........(((((((....)))))))...((((....)))).. ( -21.20) >DroYak_CAF1 129295 105 + 1 GAAGUCACGCACUCUGCCACAUUGAAAUGCAAUUAAUGCAAUAA---------------AAACGUGAGGCGCGUGAAAAAAAUAAUUUAUGGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((.....(((.(((.....((((.....))))....---------------....))).)))))))))..........((((((((((((........)))))).)))))). ( -25.02) >consensus GAAGUCACGCACUCUGCCACAUUGAAACGCAAUUAAGGCAAAAAAAAA____AA_____AAACGUGAGGCGCGUGGAAAAAAUAAUUUAUAGAAUACAAUAAACAAUGUGUUGCCAUAGU ....((((((....((((..((((.....))))...)))).......................(.....)))))))..........((((.(((((((........)))))).).)))). (-15.40 = -15.64 + 0.24)

| Location | 17,238,818 – 17,238,937 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -17.63 |
| Energy contribution | -17.39 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17238818 119 - 23771897 ACUAUGGCAACACAUUGUUUAUUGUAUUCAAUAAAUUAUUUUUUCCACGCGCCUCACGUUUUUUU-UUUACAUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUC ....((....))....((((((((....)))))))).........(((((((.....((......-...)).......((((((...((((.....))))..)))))))))))))..... ( -25.40) >DroSec_CAF1 122782 120 - 1 ACUAUGGCAACACAUUGUUUAUUGUAUUCUAUAAAUUAUUUUUUCCACGCGCAUCACGUUUUUUUAUUGAUGUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUC ..(((((.((.(((........))).)))))))............(((((((...(((((........))))).....((((((...((((.....))))..)))))))))))))..... ( -26.60) >DroSim_CAF1 116700 116 - 1 ACUAUGGCAACACAUUGUUUAUUGUAUUCUAUAAAUUAUUUUUUCCACGCGCCUCACAUUUA----UUUUUAUUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUC ..(((((.((.(((........))).)))))))............(((((((........((----....))......((((((...((((.....))))..)))))))))))))..... ( -21.40) >DroEre_CAF1 126011 105 - 1 ACUAUGGCAACACAUUGUUUAUUGUAUUCAAUAAAAUAUUUUUUCCACGCGCACCACGUUU---------------UUAUUGCAUUAAUUGCAUUUCAAUGUGGCAGAGUGCGUGACUUC ....((....)).....(((((((....)))))))..........(((((((.(((((((.---------------....((((.....))))....)))))))....)))))))..... ( -28.20) >DroYak_CAF1 129295 105 - 1 ACUAUGGCAACACAUUGUUUAUUGUAUUCCAUAAAUUAUUUUUUUCACGCGCCUCACGUUU---------------UUAUUGCAUUAAUUGCAUUUCAAUGUGGCAGAGUGCGUGACUUC ..(((((.((.(((........))).)))))))...........((((((((((((((((.---------------....((((.....))))....))))))..)).)))))))).... ( -28.20) >consensus ACUAUGGCAACACAUUGUUUAUUGUAUUCAAUAAAUUAUUUUUUCCACGCGCCUCACGUUU_____UU____UUUUUUUUUGCCUUAAUUGCGUUUCAAUGUGGCAGAGUGCGUGACUUC ..(((((.((.(((........))).)))))))............(((((((.(((((((....................(((.......)))....)))))))....)))))))..... (-17.63 = -17.39 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:20:03 2006