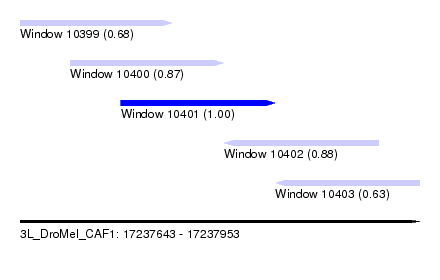
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,237,643 – 17,237,953 |
| Length | 310 |
| Max. P | 0.999812 |

| Location | 17,237,643 – 17,237,761 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.42 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.42 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237643 118 + 23771897 CAAUGAAAUUAUAUUUAUUGAAAAUGGAU-GAACACUUUUGGCCAGC-AUCGACUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAA ..(((..(((((.((((((.......)))-)))......(((((.((-(...(((((.......))))).....))).))))).)))))..)))(((((((........))))))).... ( -31.30) >DroSec_CAF1 121640 117 + 1 AAAAGAUAUU--CUUCAUCGAAAAUAUAAGGAAAAGUAUUGGCCCGC-AUCGUUUGCUUUUCUCGCAGUGUACUUGUGGGCAACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAA ....((((((--.(((..............))).)))))).((((((-(....((((.......))))......))))))).............(((((((........))))))).... ( -27.74) >DroSim_CAF1 115519 116 + 1 AACAGACAUU--CUUCAUCGAAAAUAUAU-GAAAAGUAUUGGCCAGC-AUCGCUUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAA ..........--..............(((-(.....((((((((.((-(.((((.((.......))))))....))).))))).)))....))))((((((........))))))..... ( -28.50) >DroEre_CAF1 124884 106 + 1 ------------CUUUAUUGCAAUUUUCU-GCAAUCGAUUGGCCAGC-GCCGACUGCUUUUCCCACAGUGUACUUGUGGGCCACGUAAUGGCGUAGUUGCCUCUGCCCCGGCAACUUAAA ------------....((((((......)-)))))..........((-((((..(((....(((((((.....)))))))....))).))))))(((((((........))))))).... ( -33.40) >DroYak_CAF1 128082 106 + 1 ------------UUUUAAUGAAGUUAUCU-UC-AUCUAUUGGCAAGCCAUCGACUGCUUUUGCCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAA ------------.....((((((....))-))-))....(.(((((....(.(((((.......))))))..))))).)((((.....))))..(((((((........))))))).... ( -28.80) >consensus _A__GA_AUU__CUUUAUUGAAAAUAUAU_GAAAACUAUUGGCCAGC_AUCGACUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAA .......................................(((((.((.....(((((.......)))))......)).)))))...........(((((((........))))))).... (-19.66 = -20.42 + 0.76)

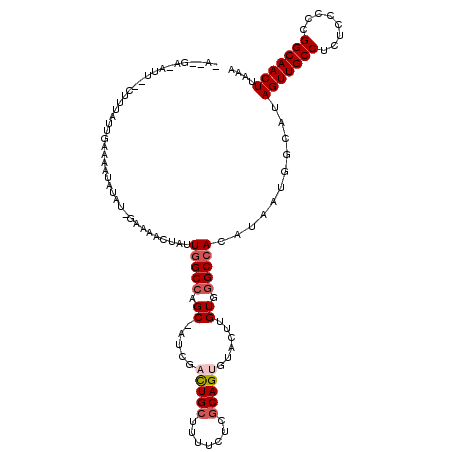
| Location | 17,237,682 – 17,237,801 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -29.36 |
| Energy contribution | -29.16 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237682 119 + 23771897 GGCCAGC-AUCGACUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCC ((((...-...((((((..((..((...((...((((((((((.....))))..(((((((........)))))))..............)))))).))..))..)).))))))..)))) ( -33.80) >DroSec_CAF1 121678 119 + 1 GGCCCGC-AUCGUUUGCUUUUCUCGCAGUGUACUUGUGGGCAACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCC (((((((-(.....))).......(((...((((((((((.(((((........(((((((........))))))).......))))).)))))).......)))).))).....))))) ( -35.97) >DroSim_CAF1 115556 119 + 1 GGCCAGC-AUCGCUUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCC (((((((-(.....))))......(((...((((..(((((((.....))))..(((((((........)))))))....................)))...)))).)))......)))) ( -33.60) >DroEre_CAF1 124911 119 + 1 GGCCAGC-GCCGACUGCUUUUCCCACAGUGUACUUGUGGGCCACGUAAUGGCGUAGUUGCCUCUGCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCC ((((.((-((((..(((....(((((((.....)))))))....))).))))))(((((((........)))))))........(((...((((......))))....))).....)))) ( -38.10) >DroYak_CAF1 128108 120 + 1 GGCAAGCCAUCGACUGCUUUUGCCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCGGGGCC .....(((.((((((((..((((((...((...((((((((((.....))))..(((((((........)))))))..............)))))).))..)))))).))))))))))). ( -43.80) >consensus GGCCAGC_AUCGACUGCUUUUCUCGCAGUGUACUUGUGGGCCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCC ((((.......((((((..((.(((...((...(((((((..((((........(((((((........))))))).......))))..))))))).))..))).)).))))))..)))) (-29.36 = -29.16 + -0.20)

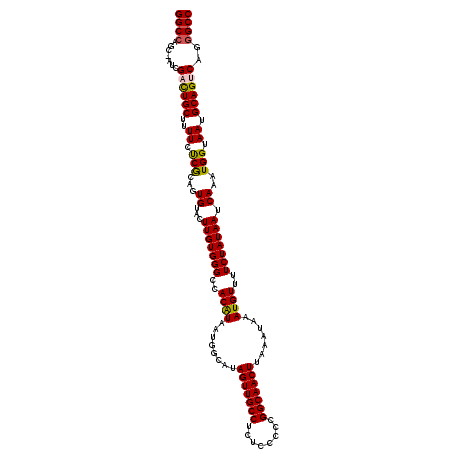
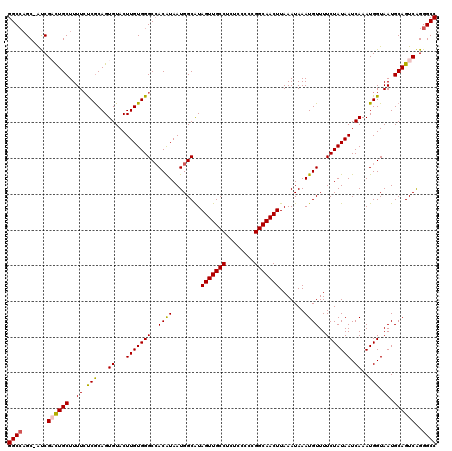
| Location | 17,237,721 – 17,237,841 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -29.02 |
| Energy contribution | -30.30 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237721 120 + 23771897 CCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCCACAGCCUCUGAACACUGCCCGAAAACCUAGCCAAGCGUUC ........((((..(((((((........))))))).........((((((....(((....)))...(((((((((((....))).))))...))))..))))))...))))....... ( -33.80) >DroSec_CAF1 121717 120 + 1 CAACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCCACAGCCUCUGCACACUGCCCGAAAACCUAGCCAAGCGUUC ........((((..(((((((........))))))).........((((((....(((....)))...(((((((((((....))).)))...)))))..))))))...))))....... ( -33.60) >DroSim_CAF1 115595 120 + 1 CCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCCACAGCCUCUGCACACUGCCCGAAAACCUAGCCAAGCGUUC ........((((..(((((((........))))))).........((((((....(((....)))...(((((((((((....))).)))...)))))..))))))...))))....... ( -33.60) >DroEre_CAF1 124950 109 + 1 CCACGUAAUGGCGUAGUUGCCUCUGCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCCGCAGCCUC-----------CGAAAACCUAGCCAAGCGUUC ..((((..((((..(((((((........))))))).........((((((....(((....)))..(((.((....)).))).....-----------.))))))...)))).)))).. ( -27.70) >DroYak_CAF1 128148 120 + 1 CCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCGGGGCCGCAGCCUCUGAACACUGCCCGAAAACCCAGCCAAGCGUUC ........((((..(((((((........))))))).........((((((....(((....)))...(((((((((((....))).))))...))))..))))))...))))....... ( -33.10) >consensus CCACAUAAUGGCAUAGUUGCCUCUCCCCCGGCAACUUAAAUAAAUGUUUUCUAUAAUCAAAUGGUAAUGCAGUCAGGGCCACAGCCUCUGAACACUGCCCGAAAACCUAGCCAAGCGUUC ........((((..(((((((........))))))).........((((((....(((....)))...(((((((((((....))).)))...)))))..))))))...))))....... (-29.02 = -30.30 + 1.28)

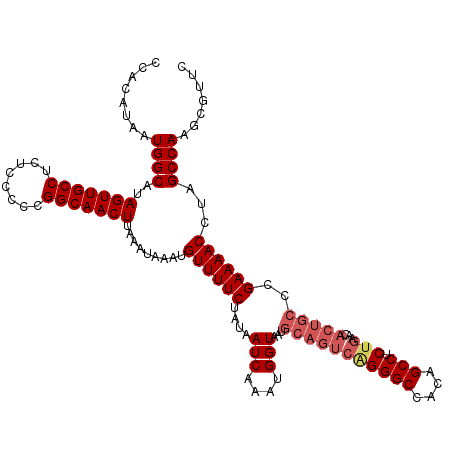
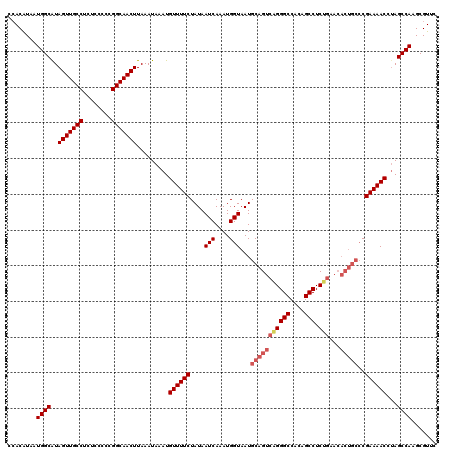
| Location | 17,237,801 – 17,237,921 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -41.62 |
| Consensus MFE | -32.74 |
| Energy contribution | -33.26 |
| Covariance contribution | 0.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237801 120 - 23771897 GGGUUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGCAAUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCUGAACGCUUGGCUAGGUUUUCGGGCAGUGUUCAGAGGCUGU ((.((((((((((((((....)))))))).(((((((..((.............))..))))))))))))).)).((((((((((((.(.((........)).)))))))))).)))... ( -44.82) >DroSec_CAF1 121797 120 - 1 GGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGAAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCUGAACGCUUGGCUAGGUUUUCGGGCAGUGUGCAGAGGCUGU (((((((((((((((((....)))))))))).(((.((((....((((......))))..))))))).)))))))((((((.(((((.(.((........)).)))))).))).)))... ( -42.50) >DroSim_CAF1 115675 120 - 1 GGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGUAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCUGAACGCUUGGCUAGGUUUUCGGGCAGUGUGCAGAGGCUGU (((((..((((((((((....)))))))))).)))))((((.((((.((((((.....)))))))))).....((((((.(((.(((......))).)))))))))........)))).. ( -43.30) >DroEre_CAF1 125030 108 - 1 GGGUUUUAGGGU-UUGGAUUUCUGAAGCCUUUGGUUCACUCGGAGUUCAGUUUAGAACGAACUAGCUAAAAGUCUGCCCUGAACGCUUGGCUAGGUUUUCG-----------GAGGCUGC (..((..(((.(-(..(....)..)).)))..))..).(((.((((((......))))...((((((((..(((......).))..))))))))....)).-----------)))..... ( -32.20) >DroYak_CAF1 128228 120 - 1 GGGCUUUAGGGUUUUGGAUUUCUGAAGCCUUCGGUUCAGUCGGAGUUCAGUUUAGAACGAACUAGCUAAAAGUCUGCCCUGAACGCUUGGCUGGGUUUUCGGGCAGUGUUCAGAGGCUGC ((((((((((((((..(....)..))))))).((((.(((....((((......))))..))))))).)))))))((((((((((((.(.((((....)))).)))))))))).)))... ( -45.30) >consensus GGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGGAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCUGAACGCUUGGCUAGGUUUUCGGGCAGUGUGCAGAGGCUGU .((((((((((((((((....))))))))))((((((..((.............))..)))))).........((((((.(((.((((....)))).)))))))))......)))))).. (-32.74 = -33.26 + 0.52)

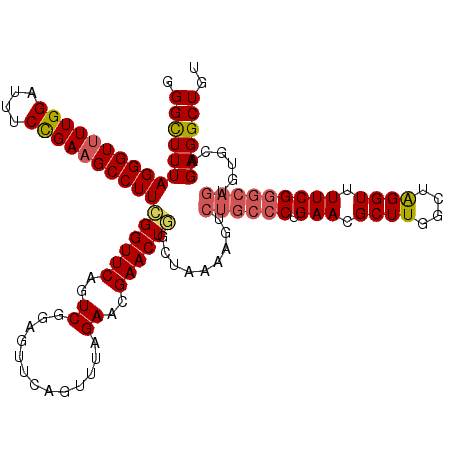
| Location | 17,237,841 – 17,237,953 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.14 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -31.38 |
| Energy contribution | -31.02 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237841 112 - 23771897 AUGAGGCUGACCUCCCUCG-UU-------CCUGGGUUCUUGGGUUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGCAAUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCU ((((((........)))))-).-------...((((....((.((((((((((((((....)))))))).(((((((..((.............))..))))))))))))).)).)))). ( -36.32) >DroSec_CAF1 121837 113 - 1 AUGAGGCUGACCUCCCUCGGUU-------CCUGGGUUCUUGGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGAAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCU ...(((..((((......))))-------)))((((....(((((((((((((((((....)))))))))).(((.((((....((((......))))..))))))).))))))))))). ( -37.60) >DroSim_CAF1 115715 113 - 1 AUGAGGCUGACCUCCCUCGGUU-------CCUGGGUUCUUGGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGUAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCU ...(((..((((......))))-------)))((((....(((((((((((((((((....)))))))))).(((.((((....((((......))))..))))))).))))))))))). ( -37.60) >DroEre_CAF1 125059 112 - 1 ACGAGGCUGACCUCCCCCGGUU-------CCUGGGUCCUUGGGUUUUAGGGU-UUGGAUUUCUGAAGCCUUUGGUUCACUCGGAGUUCAGUUUAGAACGAACUAGCUAAAAGUCUGCCCU ....(((.(((.....(..(((-------(((((((((..(((((((((((.-......)))))))))))..))...)))))).((((......))))))))..)......))).))).. ( -36.80) >DroYak_CAF1 128268 120 - 1 ACGAGGCUGACCUCCCUCGGUUUCUGGGUCCUGGGUCCUUGGGCUUUAGGGUUUUGGAUUUCUGAAGCCUUCGGUUCAGUCGGAGUUCAGUUUAGAACGAACUAGCUAAAAGUCUGCCCU ..((((((((......)))))))).((((.(((((.((..(((((((((((........)))))))))))..)))))))....((((.(((((.....)))))))))........)))). ( -39.80) >consensus AUGAGGCUGACCUCCCUCGGUU_______CCUGGGUUCUUGGGCUUUAGGGUUUUGGAUUUCCGAAGCCUUCGGUUCAGUCGGAGUUCAGUUUAGAACGAACUGGCUAAAAGUCUGCCCU .(((((........))))).............((((....(((((((((((((((((....))))))))))((((((..((.............))..))))))....))))))))))). (-31.38 = -31.02 + -0.36)

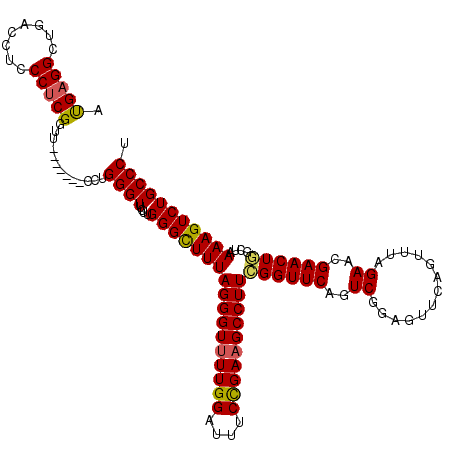
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:56 2006