
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,237,195 – 17,237,471 |
| Length | 276 |
| Max. P | 0.991596 |

| Location | 17,237,195 – 17,237,314 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -40.78 |
| Consensus MFE | -34.18 |
| Energy contribution | -33.90 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
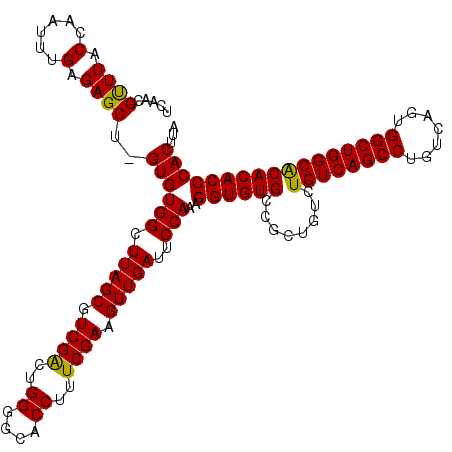
>3L_DroMel_CAF1 17237195 119 + 23771897 UCAACGUUUACCAAUUUGAGAGCU-GUGUGGCUUAGCGUCGGCUGGGGCACCUUUCGAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUA ...................(((..-(((((.......(.((((.((.((((((((.(((.....))).)))))))).)))))).)((((((((.......)))))))))))))...))). ( -44.30) >DroSec_CAF1 121188 119 + 1 UCAACGUUUACCAACUUGAGAGCU-GUGUGGCUUAGCGUCGACUGGGGCACCUUUCGAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUA ((((.(((....)))))))(((..-(((((..........((((((.((((((((.(((.....))).)))))))).)))..)))((((((((.......)))))))))))))...))). ( -42.40) >DroSim_CAF1 115033 119 + 1 UCAACGUUUACCAAUUUGAGAGCU-GUGUGGCUUAGCGUCGACUGGGGCACCUUUCGAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUA ((.(((((...........(((((-....)))))))))).)).((((((((((((.(((.....))).)))))))).....(((.((((((((.......)))))))).))))))).... ( -41.10) >DroEre_CAF1 124443 104 + 1 UCAGCGCUUACCAAUUUGAGAGCUGGUGUGGCUUAGCGUC----------------GAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCACUGGCUGGCACACACCCACUUA .((((.((((......)))).))))((((((.(((((...----------------...)))))..)))..((((((........((((((((.......)))))))))))))))))... ( -36.00) >DroYak_CAF1 127589 119 + 1 UCAACGUUUACCAAUUUGAGAGCU-GUGUGGCUUAGCGUCGGCCGGGGAACCUUUCGAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCAGUGGCUGGCGCACACCCACUUA .....((((.(......).)))).-..((((....(((((((((((.(.((((((.(((.....))).)))))).).))((((.....))))........)))))))))....))))... ( -40.10) >consensus UCAACGUUUACCAAUUUGAGAGCU_GUGUGGCUUAGCGUCGACUGGGGCACCUUUCGAAGUUGAUUCCAAAGGUGUGCCGCUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUA .....((((.(......).))))..((((((.(((((.((((..((....))..)))).)))))..)))..((((((........((((((((.......)))))))))))))))))... (-34.18 = -33.90 + -0.28)


| Location | 17,237,274 – 17,237,391 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -26.47 |
| Energy contribution | -27.24 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237274 117 + 23771897 CUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUAUCCGCCAGCUGAGCCACCCCACCA---CAAGUAGUGCACCACCGCCAGUGCACCACCGCCAGUGCACCACCCACUUGGCC .(((.((((((((.......)))))))).)))...........(((((.((.(......(((..---...((.((((((........)))))).)).....)))......))).))))). ( -33.80) >DroSec_CAF1 121267 103 + 1 CUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUAUCCGCCAGCUGAGCCACCACACCA---CCAGUAGUGCAACACCGCCAGUGC--------------ACCACCCACUUGGCC .(((.((((((((.......)))))))).)))...........(((.((((.............---.)))).).))......(((((((.--------------......))).)))). ( -28.24) >DroSim_CAF1 115112 115 + 1 CUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUAUCCGCCAGCUGAGCCACCUCACCA---CCAGCAGUGCAACACCGCCAGUGCACCAC--UCAGUGCACCACCCACUUGGCC ..........((((((..(((((((((................))))))((((....)))).))---)..)))).))......((((((((((...--...))))))........)))). ( -35.39) >DroEre_CAF1 124507 94 + 1 CUGUCUGUCAGCCUGUCACUGGCUGGCACACACCCACUUAUGCGCCAGCCGAGCCACCC-ACC-----------------------AGUGCACCAC--UCAGCGCACCACCCACUUGGCC .(((.((((((((.......)))))))).)))...............((((((......-...-----------------------.((((.....--...))))........)))))). ( -27.37) >DroYak_CAF1 127668 112 + 1 CUGUCUGUCAGCCUGUCAGUGGCUGGCGCACACCCACUUAUCCGCCAGCUGAACCACCC-ACCAGUGCCACCAGUG-----CCACCAGUGCACCAC--UCAGUGCACCACCCACUUGGCC .(((.((((((((.......)))))))).)))...........(((((.((........-....(((.((....))-----.)))..((((((...--...))))))....)).))))). ( -34.30) >consensus CUGUCUGUCAGCCUGUCAGUGGCUGGCACACACCCACUUAUCCGCCAGCUGAGCCACCCCACCA___CCAGCAGUGCA_CACCGCCAGUGCACCAC__UCAGUGCACCACCCACUUGGCC .(((.((((((((.......)))))))).)))...........(((((.((......................(((......)))..((((((........))))))....)).))))). (-26.47 = -27.24 + 0.77)


| Location | 17,237,274 – 17,237,391 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.79 |
| Mean single sequence MFE | -49.20 |
| Consensus MFE | -33.53 |
| Energy contribution | -34.00 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237274 117 - 23771897 GGCCAAGUGGGUGGUGCACUGGCGGUGGUGCACUGGCGGUGGUGCACUACUUG---UGGUGGGGUGGCUCAGCUGGCGGAUAAGUGGGUGUGUGCCAGCCACUGACAGGCUGACAGACAG ((((..(..((((((((((((.((.(((....))).)).))))))))))))..---)((((.(.((((.(((((.((......)).))).)).)))).)))))....))))......... ( -52.60) >DroSec_CAF1 121267 103 - 1 GGCCAAGUGGGUGGU--------------GCACUGGCGGUGUUGCACUACUGG---UGGUGUGGUGGCUCAGCUGGCGGAUAAGUGGGUGUGUGCCAGCCACUGACAGGCUGACAGACAG ((((.(((((.(((.--------------.((((((.(.((..((((((....---))))))..)).))))(((.((......)).))))))..))).)))))....))))......... ( -42.80) >DroSim_CAF1 115112 115 - 1 GGCCAAGUGGGUGGUGCACUGA--GUGGUGCACUGGCGGUGUUGCACUGCUGG---UGGUGAGGUGGCUCAGCUGGCGGAUAAGUGGGUGUGUGCCAGCCACUGACAGGCUGACAGACAG ((((.(((((.(((..((((((--((.(..(((..((((((...))))))..)---))......).)))))(((.((......)).))))))..))).)))))....))))......... ( -51.70) >DroEre_CAF1 124507 94 - 1 GGCCAAGUGGGUGGUGCGCUGA--GUGGUGCACU-----------------------GGU-GGGUGGCUCGGCUGGCGCAUAAGUGGGUGUGUGCCAGCCAGUGACAGGCUGACAGACAG ((((...(.(.(((((((((..--..))))))))-----------------------).)-.)((.(((.((((((((((((......))))))))))))))).)).))))......... ( -46.30) >DroYak_CAF1 127668 112 - 1 GGCCAAGUGGGUGGUGCACUGA--GUGGUGCACUGGUGG-----CACUGGUGGCACUGGU-GGGUGGUUCAGCUGGCGGAUAAGUGGGUGUGCGCCAGCCACUGACAGGCUGACAGACAG ((((.(((((.(((((((((((--((..(.(((..(((.-----((....)).)))..))-).)..)))))(((.((......)).))))))))))).)))))....))))......... ( -52.60) >consensus GGCCAAGUGGGUGGUGCACUGA__GUGGUGCACUGGCGGUG_UGCACUACUGG___UGGUGGGGUGGCUCAGCUGGCGGAUAAGUGGGUGUGUGCCAGCCACUGACAGGCUGACAGACAG ((((..((.(((((((((((......)))))))......................................(((((((.(((......))).))))))))))).)).))))......... (-33.53 = -34.00 + 0.47)



| Location | 17,237,351 – 17,237,471 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.15 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -37.03 |
| Energy contribution | -38.34 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17237351 120 - 23771897 CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCUUCGCAGUGGCCAAGUGGGUGGUGCACUGGCGGUGGUGCACUGGCGGU ........(((((((....(((((..((((.((((((....(..((((((((....)))))).))..)((((........))))..))))))..))))......)))))....))))))) ( -46.90) >DroSec_CAF1 121344 106 - 1 CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCUUCGCAGUGGCCAAGUGGGUGGU--------------GCACUGGCGGU ........(((((((....(((((.((....))((((....(..((((((((....)))))).))..)((((........))))..)))))))))--------------....))))))) ( -37.50) >DroSim_CAF1 115189 118 - 1 CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCUUCGCAGUGGCCAAGUGGGUGGUGCACUGA--GUGGUGCACUGGCGGU ........(((((((....(((((..((((.((((((....(..((((((((....)))))).))..)((((........))))..))))))..))))....--)))))....))))))) ( -47.10) >DroEre_CAF1 124569 112 - 1 CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCGCCGCAGUGGCCAAGUGGGUGGUGCGCUGA--GUGGUGCACU------ .....((((((((((....(((((.((....))((((....(..((((((((....)))))).))..)((((((....))))))..)))))))))....)).--))))))))..------ ( -42.60) >DroYak_CAF1 127743 117 - 1 CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCUCGGCAGUGGCCAAGUGGGUGGUGCACUGA--GUGGUGCACUGGUGG- .........((((((....(((((..((((.((((((....(..((((((((....)))))).))..)((((.(....).))))..))))))..))))....--)))))....))))))- ( -44.20) >consensus CAAACUGCACCGCCAACUAACCACAAUGCAAAUCCGCAGAGCAAACAAUGUCAGGUGACGUUCGUCGGGGCCUUCGCAGUGGCCAAGUGGGUGGUGCACUGA__GUGGUGCACUGGCGGU .........(((((......((((..(((......)))...(..((((((((....)))))).))..)((((........))))..))))..((((((((......))))))))))))). (-37.03 = -38.34 + 1.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:51 2006