
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,236,856 – 17,237,051 |
| Length | 195 |
| Max. P | 0.988753 |

| Location | 17,236,856 – 17,236,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -38.01 |
| Consensus MFE | -32.82 |
| Energy contribution | -32.42 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17236856 120 + 23771897 CUCAGUUGGGUUUGCCAUUAGCAUUCGCAAUGGGUUUUCGCAUACCAAUGCCCAUGUGCACUUACGAGUAGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAA (((.((.(((..(((.....)))..)(((((((((..............)))))).))).)).)))))........((((...((((((((....))))))))......))))....... ( -36.94) >DroSec_CAF1 120849 120 + 1 CUCAGUUGGGCUUGCCAUUAGCAUUCGCAAUGGGUUUUCGCAUACCAAUGCCCAUGUGCACUUACGAGUGGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAA .....((((.....(((((.((....)))))))...........(((((((((...(.((((....)))).)...))))....((((((((....))))))))...)))))))))..... ( -40.00) >DroSim_CAF1 114694 120 + 1 CUCAGUUGGGCUUGCCAUUAGCAUUCGCAAUGGGUUUUCGCAUACCAAUGCCCAUGUGCACUUACGAGUGGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAA .....((((.....(((((.((....)))))))...........(((((((((...(.((((....)))).)...))))....((((((((....))))))))...)))))))))..... ( -40.00) >DroEre_CAF1 124165 113 + 1 CUCAGUGGGGCUGGCCAUUAGCAU-UGCAAUGGAUUUUCGCAUACCAAUGCCCAUGUGCACUUA------GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCUAAGACAA ..(((.....)))(((((..((((-((..((((.....).)))..))))))..))).)).((((------(.....((((...((((((((....))))))))..))))..))))).... ( -34.60) >DroYak_CAF1 127235 114 + 1 CUCAGUUGGGUUUGCCAUUAGCAUUUGCAAUGGCUUUCCGCAUACCAAUGCCCAUGUGCACUUA------GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAA ....((((((...((((((.((....))))))))..))).....(((((((((...........------.....))))....((((((((....))))))))...)))))....))).. ( -38.49) >consensus CUCAGUUGGGCUUGCCAUUAGCAUUCGCAAUGGGUUUUCGCAUACCAAUGCCCAUGUGCACUUACGAGU_GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAA ....((..((((.(((....((((..(((.(((...........))).)))....)))).(((..........))))))....((((((((....))))))))......))))...)).. (-32.82 = -32.42 + -0.40)


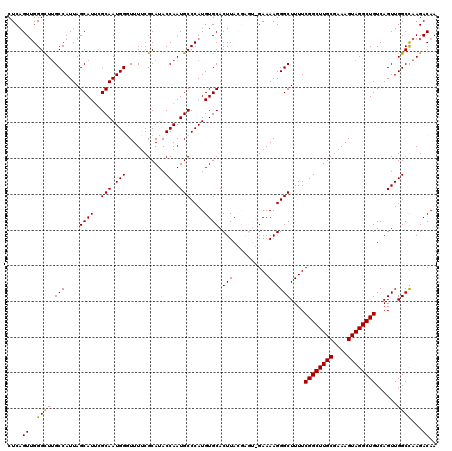
| Location | 17,236,856 – 17,236,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -29.69 |
| Energy contribution | -29.33 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17236856 120 - 23771897 UUGUCUUGGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUCUACUCGUAAGUGCACAUGGGCAUUGGUAUGCGAAAACCCAUUGCGAAUGCUAAUGGCAAACCCAACUGAG ((((((((((....((.(((.....(((((((.(((((...((((.....((((....)))).....)))).))))).))))))).....)))))...))))).)))))........... ( -32.30) >DroSec_CAF1 120849 120 - 1 UUGUCUUGGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUCCACUCGUAAGUGCACAUGGGCAUUGGUAUGCGAAAACCCAUUGCGAAUGCUAAUGGCAAGCCCAACUGAG .......(((...............(((((((.(((((...((((.....((((....)))).....)))).))))).)))))))..(((((((....)).)))))...)))........ ( -34.30) >DroSim_CAF1 114694 120 - 1 UUGUCUUGGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUCCACUCGUAAGUGCACAUGGGCAUUGGUAUGCGAAAACCCAUUGCGAAUGCUAAUGGCAAGCCCAACUGAG .......(((...............(((((((.(((((...((((.....((((....)))).....)))).))))).)))))))..(((((((....)).)))))...)))........ ( -34.30) >DroEre_CAF1 124165 113 - 1 UUGUCUUAGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUC------UAAGUGCACAUGGGCAUUGGUAUGCGAAAAUCCAUUGCA-AUGCUAAUGGCCAGCCCCACUGAG ....(((((....(((.........(((((((.(((((...((((.....------...........)))).))))).)))))))..(((((((.-..)).))))).))).....))))) ( -29.49) >DroYak_CAF1 127235 114 - 1 UUGUCUUGGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUC------UAAGUGCACAUGGGCAUUGGUAUGCGGAAAGCCAUUGCAAAUGCUAAUGGCAAACCCAACUGAG (((((..(.....).))))).....(((((((.(((((...((((.....------...........)))).))))).))))))).((((((((....)).))))))............. ( -33.89) >consensus UUGUCUUGGCCAACUGACAGCCUACUUUCGCAAGCCGAAAAGCCCUUUUC_ACUCGUAAGUGCACAUGGGCAUUGGUAUGCGAAAACCCAUUGCGAAUGCUAAUGGCAAGCCCAACUGAG ((((((((((....((.(((.....(((((((.(((((...((((......................)))).))))).))))))).....)))))...))))).)))))........... (-29.69 = -29.33 + -0.36)


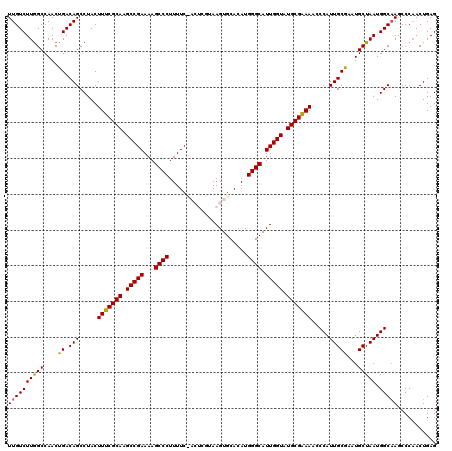
| Location | 17,236,896 – 17,237,013 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.54 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -33.51 |
| Energy contribution | -33.35 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17236896 117 + 23771897 CAUACCAAUGCCCAUGUGCACUUACGAGUAGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGAAGA---UGAUGAAGGGGAG ..........(((.....(((((...(((......(((((((.((((((((....))))))))......(((((.......)))))))))))).)))...))).---))......))).. ( -36.80) >DroSec_CAF1 120889 117 + 1 CAUACCAAUGCCCAUGUGCACUUACGAGUGGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA---UGAUGAAGGGGAG ....((...((((...(.((((....)))).)...))))((((((((((((....)))))))(((..(((((((((.(........)...))))))))))))..---....))))))).. ( -41.80) >DroSim_CAF1 114734 117 + 1 CAUACCAAUGCCCAUGUGCACUUACGAGUGGAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA---UGAUGAAGGGGAG ....((...((((...(.((((....)))).)...))))((((((((((((....)))))))(((..(((((((((.(........)...))))))))))))..---....))))))).. ( -41.80) >DroEre_CAF1 124204 111 + 1 CAUACCAAUGCCCAUGUGCACUUA------GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCUAAGACAAAUGGCCGCAGCUUGGCUAAGACGA---UGAUGCAGAGGAG ....((..(((.(((((...((((------(..(((((((...((((((((....))))))))..))))(((((.......)))))....)))..))))))).)---))..)))..)).. ( -36.00) >DroYak_CAF1 127275 114 + 1 CAUACCAAUGCCCAUGUGCACUUA------GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGAGGAUGAUGAGGAGGAG (((.((..(((......)))((((------(....(((((((.((((((((....))))))))......(((((.......))))))))))))..)))))....)))))........... ( -38.00) >consensus CAUACCAAUGCCCAUGUGCACUUACGAGU_GAAAAGGGCUUUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA___UGAUGAAGGGGAG ....((...((((......................))))((((((((((((....)))))))(((..(((((((((.(........)...)))))))))))).........))))))).. (-33.51 = -33.35 + -0.16)

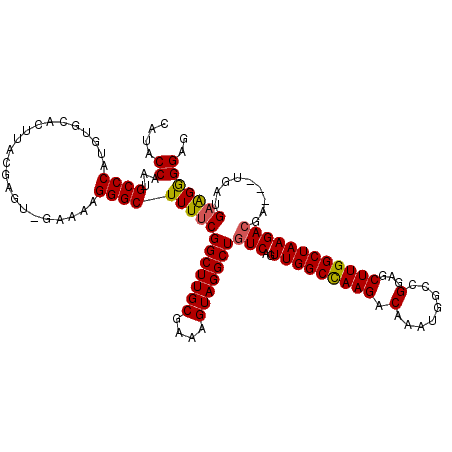

| Location | 17,236,936 – 17,237,051 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -38.06 |
| Consensus MFE | -35.90 |
| Energy contribution | -36.14 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17236936 115 + 23771897 UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGAAGA---UGAUGAAGGGGAGAAGCCGUUAACACA--UGCUCAGACGCGACGAGUGACGGG (((((((((((....)))))))((((.(((((((((.(........)...))))))))).....---))))......))))..((((((.....--(((......))).....)))))). ( -36.40) >DroSec_CAF1 120929 115 + 1 UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA---UGAUGAAGGGGAGAAGCCGUUAACACA--UGCUCAGACGCGACGACUGACGGG ....(((((((....)))))))((((((((((((.......))))).((((..((((....(..---(.....)..)....))))((....)).--.)))).........)))))))... ( -38.70) >DroSim_CAF1 114774 115 + 1 UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA---UGAUGAAGGGGAGAAGCCGUUAACACA--UGCUCAGACGCGACGACUGACGGG ....(((((((....)))))))((((((((((((.......))))).((((..((((....(..---(.....)..)....))))((....)).--.)))).........)))))))... ( -38.70) >DroEre_CAF1 124238 115 + 1 UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCUAAGACAAAUGGCCGCAGCUUGGCUAAGACGA---UGAUGCAGAGGAGGAGCCGUUAACACA--UGCUCAGACGCGACGACUGACGGG ....(((((((....)))))))((((((((((((.......))))(((..(((.((........---....)).)))...((((.((....)).--.))))....))).))))))))... ( -37.30) >DroYak_CAF1 127309 120 + 1 UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGAGGAUGAUGAGGAGGAGAAGCCGUUAACACAUAUGCUCAGACGCGACGACUGACGGG ....(((((((....)))))))((((((((((((((.(........)...))))))......(((.(((.((.(..((.....)).....).))))).)))........))))))))... ( -39.20) >consensus UUUCGGCUUGCGAAAGUAGGCUGUCAGUUGGCCAAGACAAAUGGCCGGAGCUUGGCUAAGACGA___UGAUGAAGGGGAGAAGCCGUUAACACA__UGCUCAGACGCGACGACUGACGGG ....(((((((....)))))))((((((((((((.......))))).((((..((((........................))))((....))....)))).........)))))))... (-35.90 = -36.14 + 0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:47 2006