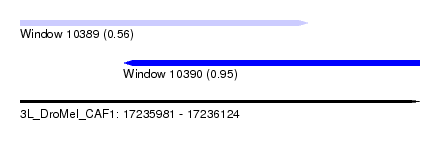
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,235,981 – 17,236,124 |
| Length | 143 |
| Max. P | 0.948059 |

| Location | 17,235,981 – 17,236,084 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -20.76 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17235981 103 + 23771897 ---UCCCCAUCUUUAUUUCCACAGGCCGAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCUGCUU--------------CUCGCCUCUCGGCUCAUGCAACAGCACUUUGGCCAAGUGUC ---................(((((((.((((((((.(((.(....((...))....)))))))))--------------)))))))...((((..(((....)))....))))..))).. ( -30.60) >DroSec_CAF1 119996 103 + 1 ---UCCCCAUCUUUAUUUCCACAGGCCGAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCCGCUU--------------CUCGCCUCUCGGCUCGUGCAACAGCACUUUGGCCAAGUGUC ---................(((((((.(((((((..(((.(....((...))....)))).))))--------------)))))))...((((.((((....))))...))))..))).. ( -31.50) >DroSim_CAF1 113820 103 + 1 ---UCCCCAUCUUCAUUUCCACAGGCCGAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCCGCUU--------------CUCGCCUCUCGGCUCGUGCAACAGCACUUUGGCCAAGUGUC ---................(((((((.(((((((..(((.(....((...))....)))).))))--------------)))))))...((((.((((....))))...))))..))).. ( -31.50) >DroEre_CAF1 123386 114 + 1 ------CCACUUGCAUUUCCCCUGGCCCAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCUGCUUUUCGCUGUUUGCUUCUCGCCUCACGGCUCCUGCAACAGCACUUUGGCCAAAUGUC ------......(((((.....(((((.(((((((.(((.(....((...))....))))))))))).((((((.((.....(((....)))....)))))))).....)))))))))). ( -32.30) >DroYak_CAF1 126340 115 + 1 CAAUCCCCAUUUCCAUUUCCAACGGCCGAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCUGCUUUUCACUUUUCACGUCUCGCCUC-----UCAUGCAACAACACUUUGGCCAAGUGCC .......................(((((((..((.((((((...((((.((.......))........((.......))...)))).-----.))))))...))..)))))))....... ( -22.50) >consensus ___UCCCCAUCUUCAUUUCCACAGGCCGAGAAGUAUGCGUGUUCAGGCUGCCAUUUCCGCUGCUU______________CUCGCCUCUCGGCUCAUGCAACAGCACUUUGGCCAAGUGUC .......................(((((((..((.((((((...((((.((.......)).)))).................(((....))).))))))...))..)))))))....... (-20.76 = -21.44 + 0.68)


| Location | 17,236,018 – 17,236,124 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.15 |
| Mean single sequence MFE | -44.08 |
| Consensus MFE | -36.00 |
| Energy contribution | -36.48 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17236018 106 - 23771897 UGGCAAUGAGCUGCCGUGAGCCAUGGGCGUUUGGAGCAGCGACACUUGGCCAAAGUGCUGUUGCAUGAGCCGAGAGGCGAG--------------AAGCAGCGGAAAUGGCAGCCUGAAC .........((((((((...((....((.......((((((.(((((.....))))).))))))....(((....)))...--------------..))...))..))))))))...... ( -40.80) >DroSec_CAF1 120033 106 - 1 UGGCAAUGAGCUGCCGUGAGCCAUUGGCGUUUGGAGCAGCGACACUUGGCCAAAGUGCUGUUGCACGAGCCGAGAGGCGAG--------------AAGCGGCGGAAAUGGCAGCCUGAAC .........((((((((...((...(.(((((...((((((.(((((.....))))).)))))).(..(((....)))..)--------------))))).)))..))))))))...... ( -43.40) >DroSim_CAF1 113857 106 - 1 UGGCAAUGAGCUGCCGUGAGCCAUUGGCGUUUGGAGCAGCGACACUUGGCCAAAGUGCUGUUGCACGAGCCGAGAGGCGAG--------------AAGCGGCGGAAAUGGCAGCCUGAAC .........((((((((...((...(.(((((...((((((.(((((.....))))).)))))).(..(((....)))..)--------------))))).)))..))))))))...... ( -43.40) >DroEre_CAF1 123420 120 - 1 UGGCAAUGGGCUGCCGUGAGGCAUUGGCGCUCCGAGCAGCGACAUUUGGCCAAAGUGCUGUUGCAGGAGCCGUGAGGCGAGAAGCAAACAGCGAAAAGCAGCGGAAAUGGCAGCCUGAAC .(((((((.(((((((.(((((....)).))))).)))))..))))..((((.....(((((((....(((....))).....((.....)).....)))))))...)))).)))..... ( -50.40) >DroYak_CAF1 126380 115 - 1 UGGCAAUGAGCUGCCGUGAGCCAUUGGCGCUCCGAGCGGCGGCACUUGGCCAAAGUGUUGUUGCAUGA-----GAGGCGAGACGUGAAAAGUGAAAAGCAGCGGAAAUGGCAGCCUGAAC .........((((((((...((.(((((.(((((.((((((((((((.....)))))))))))).)).-----)))))...((.......))......))).))..))))))))...... ( -42.40) >consensus UGGCAAUGAGCUGCCGUGAGCCAUUGGCGUUUGGAGCAGCGACACUUGGCCAAAGUGCUGUUGCACGAGCCGAGAGGCGAG______________AAGCAGCGGAAAUGGCAGCCUGAAC .........((((((((...((....((.......(((((..(((((.....)))))..)))))....(((....)))...................))...))..))))))))...... (-36.00 = -36.48 + 0.48)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:43 2006