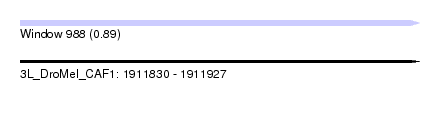
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,911,830 – 1,911,927 |
| Length | 97 |
| Max. P | 0.894440 |

| Location | 1,911,830 – 1,911,927 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -20.00 |
| Energy contribution | -20.17 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1911830 97 + 23771897 ----CCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCGCCC--------------GAAAACCG-A--AGCG ----((((.(((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))))(((.(--------------(.....))-.--.))) ( -28.60) >DroVir_CAF1 40345 95 + 1 ----UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAUU-CUUUUUUUA-----------------AAAACGC-GCGCGCG ----...((((...))))((((((((((((((..(((((((......)))))))....)))...))))))))))).-.........-----------------....(((-....))) ( -24.10) >DroGri_CAF1 39555 110 + 1 ----UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAA---UCUGCACCGCCAAAGGGUCACUUAAAAACGC-GACAGCG ----..(((((((((....(((((((((((((..(((((((......)))))))....)))...))))))))))..---...((...))....))))))))).....(((-....))) ( -31.80) >DroWil_CAF1 51216 96 + 1 ----UCAGGUGAUUCAUCAUUUCAAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUUGAAGGCUC------------------AAACCGCGACCGCG ----...(((..((((...((((.((((((((..(((((((......)))))))....)))...))))).))))...)))).....------------------..)))((....)). ( -21.10) >DroAna_CAF1 38443 97 + 1 ----UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAGUCGCCGGGCCUC--------------AAAACCGC-G--UGCG ----...(((((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))((.....--------------....))..-.--.... ( -29.00) >DroPer_CAF1 39566 98 + 1 GGAUUCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGC---C--------------GAAACCGC-G--UGCG ((.(((.(((((((.....(((((((((((((..(((((((......)))))))....)))...)))))))))))))))))...---.--------------))).))..-.--.... ( -28.90) >consensus ____UCAGGUGAUUCAUCAUUUCGAGUGUUGUUCAUUGCAACGUUUAUUGCGAUUUUUACAAACGCACUCGAAAAAUCGCUGGCUC_C_______________AAAACGC_G__AGCG .......((((...)))).(((((((((((((..(((((((......)))))))....)))...))))))))))............................................ (-20.00 = -20.17 + 0.17)

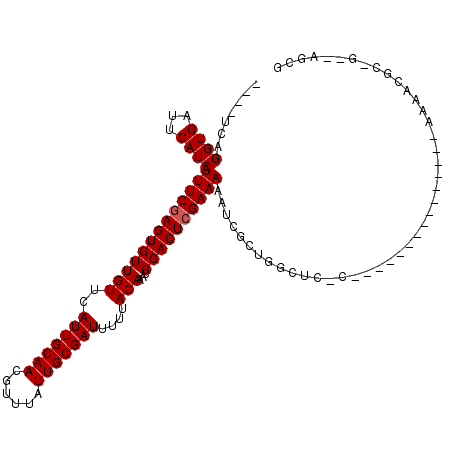
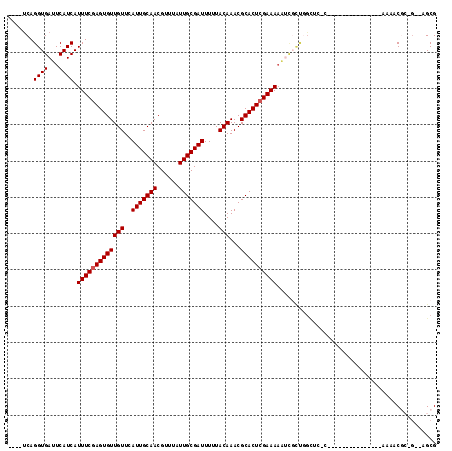
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:31 2006