
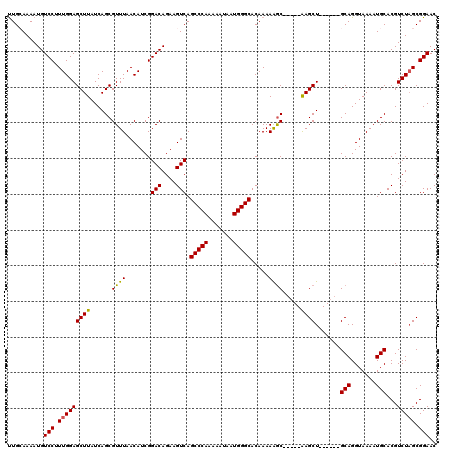
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,230,637 – 17,231,232 |
| Length | 595 |
| Max. P | 0.999945 |

| Location | 17,230,637 – 17,230,746 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -24.66 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230637 109 - 23771897 UUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC-----AAGCU------GCAGGUAAAAUGCACGUCUAGCGGAAC ((((....(((((..((..(((....)))......))..))))).......(((((.......))))).......))-----))(((------(.(.((.......)).).))))..... ( -26.30) >DroSec_CAF1 114657 109 - 1 UUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC-----AAGCU------GCAGGUAAAAUGCACGUCUAGCGGAAC ((((....(((((..((..(((....)))......))..))))).......(((((.......))))).......))-----))(((------(.(.((.......)).).))))..... ( -26.30) >DroSim_CAF1 108443 109 - 1 UUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAGAC-----AAGCU------GCAGGUAAAAUGCACGUCUAGCGGAAC ((((....(((((..((..(((....)))......))..))))).......(((((.......))))).....((((-----....(------(((.......)))).)))).))))... ( -26.50) >DroEre_CAF1 118002 108 - 1 CUGGAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC-----GAGCU------GCAGGUAAAAUGCACGUCU-GCGGAAC .(((....(((((..((..(((....)))......))..)))))....)))(((((.......))))).......((-----(..((------(((.......)))).)..)-))..... ( -28.90) >DroYak_CAF1 120810 120 - 1 UUGGAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGCUGCACAAGCUGGUAAAGCAGGUAAAAUGCACGUCUAGCGGAAC ..........(((..((.(((((.................(((....))).(((((.......))))).....))))).))...(((((....(((.......)))....)))))))).. ( -31.00) >consensus UUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC_____AAGCU______GCAGGUAAAAUGCACGUCUAGCGGAAC ..........(((.(((((((((......((((.......(((....))).(((((.......))))).....)))).....)))).......(((.......)))...))))).))).. (-24.66 = -24.38 + -0.28)


| Location | 17,230,669 – 17,230,786 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.46 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230669 117 - 23771897 UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGUUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC--- ((((....))))(((...(((.((...............)).)))...(((((..((..(((....)))......))..)))))...))).(((((.......))))).........--- ( -28.96) >DroSec_CAF1 114689 117 - 1 UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGUUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC--- ((((....))))(((...(((.((...............)).)))...(((((..((..(((....)))......))..)))))...))).(((((.......))))).........--- ( -28.96) >DroSim_CAF1 108475 117 - 1 UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGUUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAGAC--- ((((....))))(((...(((.((...............)).)))...(((((..((..(((....)))......))..)))))...))).(((((.......))))).........--- ( -28.96) >DroEre_CAF1 118033 117 - 1 UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGCUGGAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC--- ((((....))))(((............((((........)))).....(((((..((..(((....)))......))..)))))...))).(((((.......))))).........--- ( -31.90) >DroYak_CAF1 120850 120 - 1 UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGUUGGAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGCUGC ((((....))))...............(((((.........(((....(((((..((..(((....)))......))..)))))....)))(((((.......)))))......))))). ( -30.80) >consensus UGCCACUCGGCAGACAAGUGCUACUUACGGCUAUUUUAUGUUGCAAAAUGUCCUUUGGAGCUUAUCAGCGUUUAACAUCGGACAGAAGUCAGCCCAAAAAUAAUGGGCACAAAAAGC___ ((((....))))(((............((((........)))).....(((((..((..(((....)))......))..)))))...))).(((((.......)))))............ (-29.52 = -29.36 + -0.16)



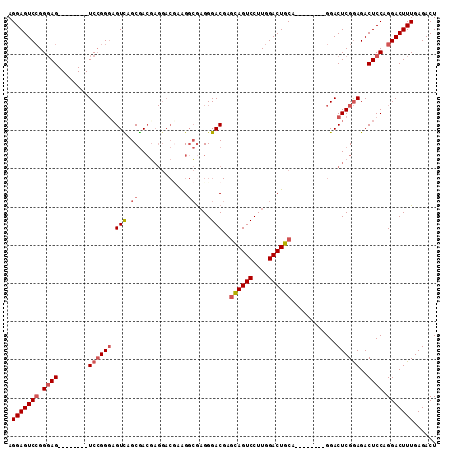
| Location | 17,230,786 – 17,230,882 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -42.75 |
| Consensus MFE | -25.90 |
| Energy contribution | -27.27 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230786 96 + 23771897 AGGAGUCCGGGAG--------UCCGGGCGUCAGCGACGAGGACGAAGGCGAGGGACGAGCAGUCCUUGGACUGCA--------GGACUCGGAGACUCCAGGACUUUGAGACU .(((((((.((((--------(((..((....))..((((..(...(((.((((((.....)))))).).))...--------)..))))).)))))).)))))))...... ( -43.30) >DroSec_CAF1 114806 96 + 1 AGGAGUCCGGGAG--------UCCGGGAGUCAGCGAAGAGGACGAAGGCGAGGGACGAGCAGUCCUUGGACUGCA--------GGACUCGGAGACUCCAGGACUUUGAGACU ...((((..((((--------(((.((((((......(((..(...(((.((((((.....)))))).).))...--------)..)))...)))))).)))))))..)))) ( -42.80) >DroSim_CAF1 108592 104 + 1 AGGAGUCCGGGAGUCGAGGUGUCCGGGAGUUAGCGACGAGGACUAAGGCAAGGGACGAGCAGUCCUUGGACUGCA--------GGACUCGGAGACUCCAGGACUUUGAGACU .(((((((.((((((..((.((((.(.((((.....((((((((...((.........)))))))))))))).).--------))))))...)))))).)))))))...... ( -43.20) >DroEre_CAF1 118150 94 + 1 AGGAGUCGGG-AG--------UC-GGCAGUU-GCAACUC----CGGGCAGCGGGAC---GAGUCCUUGGACUCCAGGACUCCAGGACUCGAAGACUCCAGGACUUUGAGACU .((((((.((-((--------((-....(((-((.....----...)))))....(---(((((((.(((.((...)).)))))))))))..))))))..))))))...... ( -41.70) >consensus AGGAGUCCGGGAG________UCCGGGAGUCAGCGACGAGGACGAAGGCGAGGGACGAGCAGUCCUUGGACUGCA________GGACUCGGAGACUCCAGGACUUUGAGACU .(((((((.((((........((((((.(((.((.............))....)))..((((((....))))))............))))))..)))).)))))))...... (-25.90 = -27.27 + 1.37)

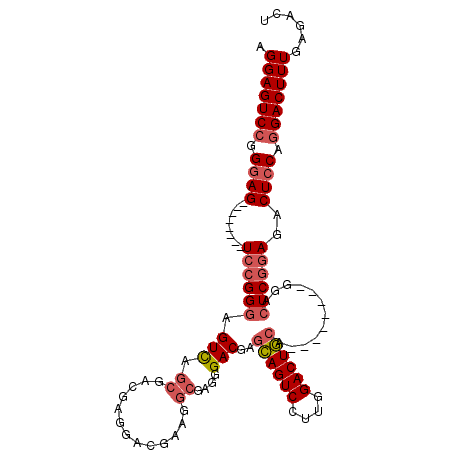
| Location | 17,230,786 – 17,230,882 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230786 96 - 23771897 AGUCUCAAAGUCCUGGAGUCUCCGAGUCC--------UGCAGUCCAAGGACUGCUCGUCCCUCGCCUUCGUCCUCGUCGCUGACGCCCGGA--------CUCCCGGACUCCU ..............(((((((..((((((--------.((((((....)))))).((((....((...((....))..)).))))...)))--------)))..))))))). ( -37.80) >DroSec_CAF1 114806 96 - 1 AGUCUCAAAGUCCUGGAGUCUCCGAGUCC--------UGCAGUCCAAGGACUGCUCGUCCCUCGCCUUCGUCCUCUUCGCUGACUCCCGGA--------CUCCCGGACUCCU (((((...(((((.((((((..((((...--------.((((((....)))))).((.....))...........))))..)))))).)))--------))...)))))... ( -37.20) >DroSim_CAF1 108592 104 - 1 AGUCUCAAAGUCCUGGAGUCUCCGAGUCC--------UGCAGUCCAAGGACUGCUCGUCCCUUGCCUUAGUCCUCGUCGCUAACUCCCGGACACCUCGACUCCCGGACUCCU ........(((((.((((((...(((...--------.((((((....))))))..((((......(((((.......))))).....))))..))))))))).)))))... ( -37.30) >DroEre_CAF1 118150 94 - 1 AGUCUCAAAGUCCUGGAGUCUUCGAGUCCUGGAGUCCUGGAGUCCAAGGACUC---GUCCCGCUGCCCG----GAGUUGC-AACUGCC-GA--------CU-CCCGACUCCU ..............((((((..(((((((((((.((...)).))).)))))))---)...........(----(((((((-....).)-))--------))-)).)))))). ( -38.80) >consensus AGUCUCAAAGUCCUGGAGUCUCCGAGUCC________UGCAGUCCAAGGACUGCUCGUCCCUCGCCUUCGUCCUCGUCGCUAACUCCCGGA________CUCCCGGACUCCU ..............(((((((..(((............((((((....))))))......................(((........))).........)))..))))))). (-20.10 = -20.72 + 0.62)

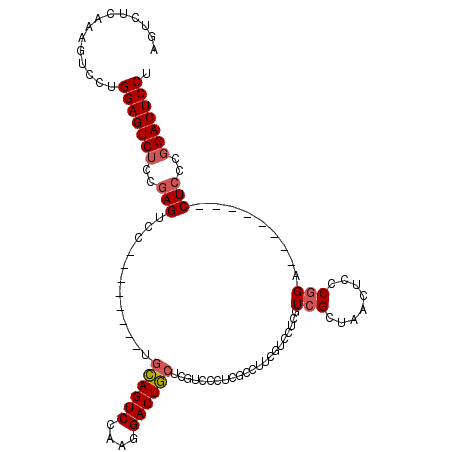

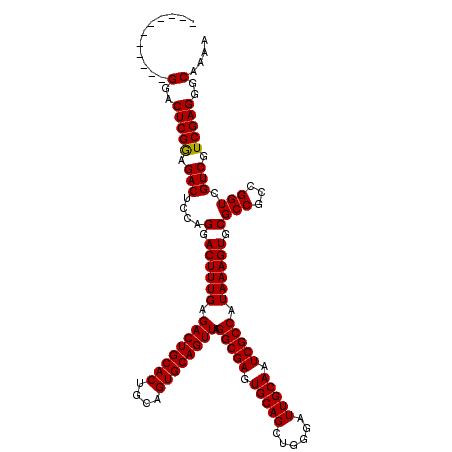
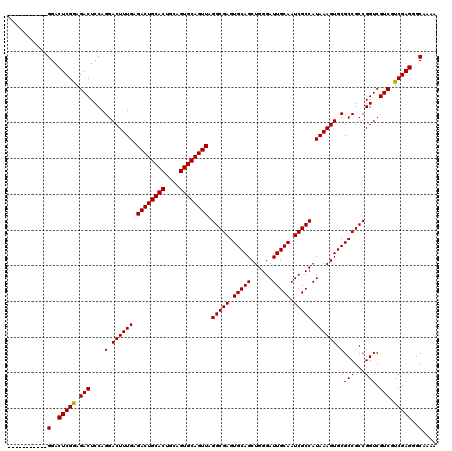
| Location | 17,230,853 – 17,230,962 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -45.24 |
| Consensus MFE | -44.68 |
| Energy contribution | -44.44 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230853 109 + 23771897 -----------GGACUCGGAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA -----------(..((((..(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).)))..))))..).... ( -44.90) >DroSec_CAF1 114873 109 + 1 -----------GGACUCGGAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA -----------(..((((..(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).)))..))))..).... ( -44.90) >DroSim_CAF1 108667 109 + 1 -----------GGACUCGGAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA -----------(..((((..(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).)))..))))..).... ( -44.90) >DroEre_CAF1 118212 112 + 1 C--------CAGGACUCGAAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA .--------..(..(((((.(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).))).)))))..).... ( -45.10) >DroYak_CAF1 121002 120 + 1 CGGUGACUACAGGACUCGAAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA .(....)....(..(((((.(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).))).)))))..).... ( -46.40) >consensus ___________GGACUCGGAGACUCCAGGACUUUGAGACUGCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAA ...........(..(((((.(((....(.((((((.((((((((....)))))))).(((((.(((((......))))).))))).)))))).)(((...))).))).)))))..).... (-44.68 = -44.44 + -0.24)

| Location | 17,230,853 – 17,230,962 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -40.94 |
| Energy contribution | -40.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.75 |
| SVM RNA-class probability | 0.999945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230853 109 - 23771897 UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUCCGAGUCC----------- .......((((..(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).)))..))))...----------- ( -41.30) >DroSec_CAF1 114873 109 - 1 UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUCCGAGUCC----------- .......((((..(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).)))..))))...----------- ( -41.30) >DroSim_CAF1 108667 109 - 1 UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUCCGAGUCC----------- .......((((..(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).)))..))))...----------- ( -41.30) >DroEre_CAF1 118212 112 - 1 UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUUCGAGUCCUG--------G ....(..(((((.(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).))).)))))..)..--------. ( -44.00) >DroYak_CAF1 121002 120 - 1 UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUUCGAGUCCUGUAGUCACCG ....(..(((((.(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).))).)))))..)........... ( -44.00) >consensus UUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGCAGUCUCAAAGUCCUGGAGUCUCCGAGUCC___________ .......((((..(((..((((.(((.........(((((.((((........)))).)))))..(((((((....)))))))......))))))).)))..)))).............. (-40.94 = -40.94 + 0.00)

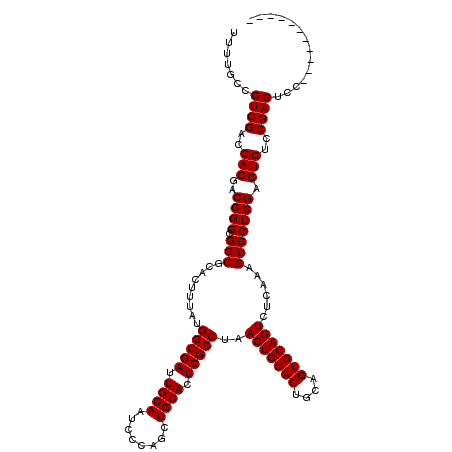
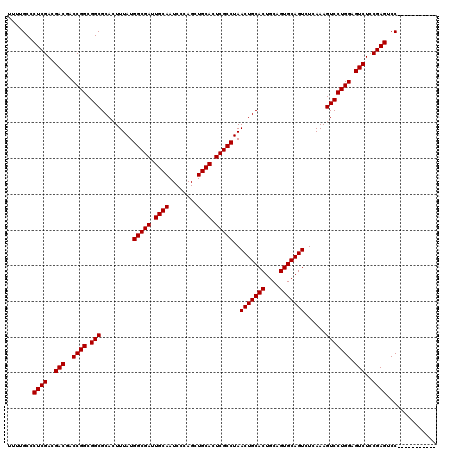
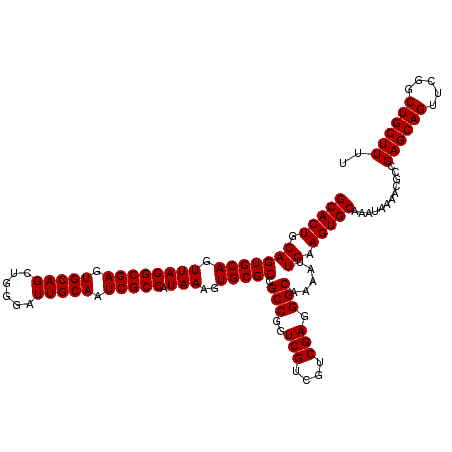
| Location | 17,230,882 – 17,231,002 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -43.60 |
| Energy contribution | -43.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17230882 120 + 23771897 GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUU (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. ( -43.60) >DroSec_CAF1 114902 120 + 1 GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUU (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. ( -43.60) >DroSim_CAF1 108696 120 + 1 GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUU (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. ( -43.60) >DroEre_CAF1 118244 120 + 1 GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGGCGAGCAGUUCGGCUGCUUUG (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. ( -43.60) >DroYak_CAF1 121042 120 + 1 GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUG (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. ( -43.60) >consensus GCACUGCAGUGCAGUUAGGCGAGUGCAGCUGGGAUUGCAAUCGCCAUAAAGUGCGCCGCCGGUCGUCGUCGAGGGCAAAAUUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUU (((((.(((((((.((((((((.(((((......))))).))))).)))..))))).(((..(((....))).))).....)).)))))............((((((.....)))))).. (-43.60 = -43.60 + 0.00)


| Location | 17,230,882 – 17,231,002 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -40.12 |
| Consensus MFE | -39.56 |
| Energy contribution | -39.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
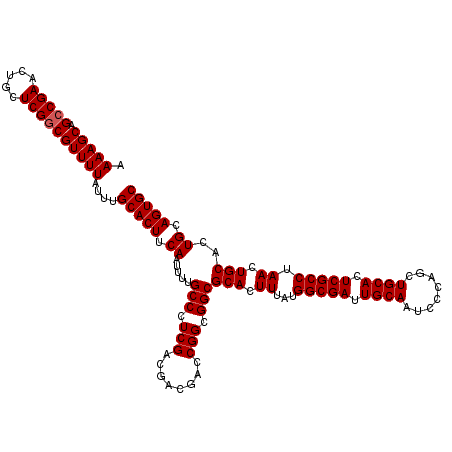
>3L_DroMel_CAF1 17230882 120 - 23771897 AAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC .(((((.(((((.....))))))))))....(((((.((.....(((.(((........))).)))(((.((...(((((.((((........)))).))))).)).)))..)).))))) ( -41.10) >DroSec_CAF1 114902 120 - 1 AAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC .(((((.(((((.....))))))))))....(((((.((.....(((.(((........))).)))(((.((...(((((.((((........)))).))))).)).)))..)).))))) ( -41.10) >DroSim_CAF1 108696 120 - 1 AAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC .(((((.(((((.....))))))))))....(((((.((.....(((.(((........))).)))(((.((...(((((.((((........)))).))))).)).)))..)).))))) ( -41.10) >DroEre_CAF1 118244 120 - 1 CAAAGCAGCCGAACUGCUCGCCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC ....(((((((..(((.(((.((((......(((.........)))....)))).))).)))))))(((.((...(((((.((((........)))).))))).)).)))..)))..... ( -36.20) >DroYak_CAF1 121042 120 - 1 CAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC .(((((.(((((.....))))))))))....(((((.((.....(((.(((........))).)))(((.((...(((((.((((........)))).))))).)).)))..)).))))) ( -41.10) >consensus AAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAAUUUUGCCCUCGACGACGACCGGCGGCGCACUUUAUGGCGAUUGCAAUCCCAGCUGCACUCGCCUAACUGCACUGCAGUGC .(((((.(((((.....))))))))))....(((((.((.....(((.(((........))).)))(((.((...(((((.((((........)))).))))).)).)))..)).))))) (-39.56 = -39.76 + 0.20)


| Location | 17,230,962 – 17,231,081 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.45 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -28.57 |
| Energy contribution | -29.57 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
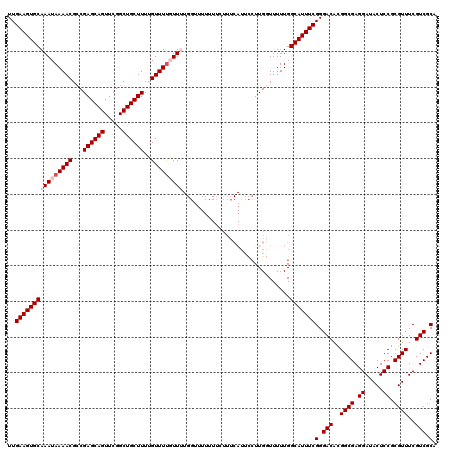
>3L_DroMel_CAF1 17230962 119 + 23771897 UUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUUGUUUUGUUUUGGUUUUUUU-UUUCAUUCCUUGGUUUUUGGCAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCA ..(((((((((((((((((..((((((.....)))))).))))))))))..........-..(((.....)))......)))))))(.(((..((((.((.....)).))))..))).). ( -34.50) >DroSec_CAF1 114982 120 + 1 UUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUUGUUUUGUUUUGGUUUUUUUCUUUCAUUCCUUGGUUUUUGGCAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCA ..(((((((((((((((((..((((((.....)))))).)))))))))).............(((.....)))......)))))))(.(((..((((.((.....)).))))..))).). ( -34.50) >DroSim_CAF1 108776 120 + 1 UUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUUGUUUUGUUUUGGUUUUUUUCUUUCAUUCCUUGGUUUUUGGCAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCA ..(((((((((((((((((..((((((.....)))))).)))))))))).............(((.....)))......)))))))(.(((..((((.((.....)).))))..))).). ( -34.50) >DroEre_CAF1 118324 99 + 1 UUGAAGUGCAAAUAAAACGGCGAGCAGUUCGGCUGCUUUGGUUUCUUU---------------------UUGUUUUUUGGCAUUUCGGGACGUGGCGAGGAUACUCCGCGUUUCGUCGCA ..(((((((((((((((.(((((((((.....))))))..)))...))---------------------))))))....)))))))(.((((..(((.((.....)).)))..)))).). ( -33.90) >DroYak_CAF1 121122 108 + 1 UUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUGGUUUUUUU----------UCUUU--UUUCUUGUUUUUUGGCAUUUCGGGACAUGGCGAGGAUACUCCGCGUUUCGUCGCA ..(((((((....((((((((((((((.....)))).)))))......----------.....--......)))))...)))))))(.(((..((((.((.....)).))))..))).). ( -32.92) >consensus UUGAAGUGCAAAUAAAACGCCGAGCAGUUCGGCUGCUUUUGUUUUGUUUUGGUUUUUUUCUUUCAUUCCUUGGUUUUUGGCAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCA ..((((((((((((((((...((((((.....))))))..)))))))))..............................)))))))(.(((..((((.((.....)).))))..))).). (-28.57 = -29.57 + 1.00)


| Location | 17,230,962 – 17,231,081 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.45 |
| Mean single sequence MFE | -24.87 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.70 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
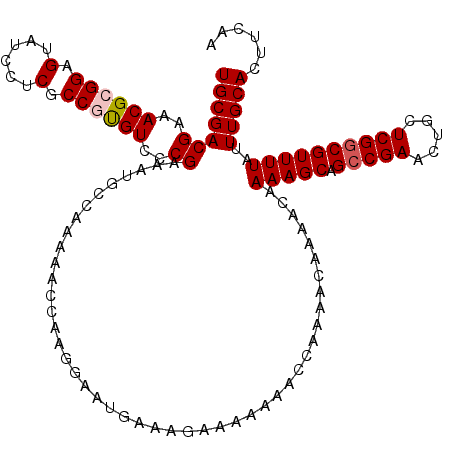
>3L_DroMel_CAF1 17230962 119 - 23771897 UGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUGCCAAAAACCAAGGAAUGAAA-AAAAAAACCAAAACAAAACAAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAA (((((((..((((((.(......).))))))..))(((((((.........((.......-.......)).............(((((.....))))).)))))))...)))))...... ( -27.64) >DroSec_CAF1 114982 120 - 1 UGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUGCCAAAAACCAAGGAAUGAAAGAAAAAAACCAAAACAAAACAAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAA (((((((..((((((.(......).))))))..))(((((((.........((...............)).............(((((.....))))).)))))))...)))))...... ( -27.56) >DroSim_CAF1 108776 120 - 1 UGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUGCCAAAAACCAAGGAAUGAAAGAAAAAAACCAAAACAAAACAAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAA (((((((..((((((.(......).))))))..))(((((((.........((...............)).............(((((.....))))).)))))))...)))))...... ( -27.56) >DroEre_CAF1 118324 99 - 1 UGCGACGAAACGCGGAGUAUCCUCGCCACGUCCCGAAAUGCCAAAAAACAA---------------------AAAGAAACCAAAGCAGCCGAACUGCUCGCCGUUUUAUUUGCACUUCAA .(.((((...((.((.....)).))...)))).)(((.(((.(((((((..---------------------...(....)..(((((.....)))))....))))).)).))).))).. ( -19.60) >DroYak_CAF1 121122 108 - 1 UGCGACGAAACGCGGAGUAUCCUCGCCAUGUCCCGAAAUGCCAAAAAACAAGAAA--AAAGA----------AAAAAAACCAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAA (((((.(((((((.((((((..(((........))).))))..............--.....----------............((((.....)))))).)))))))..)))))...... ( -22.00) >consensus UGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUGCCAAAAACCAAGGAAUGAAAGAAAAAAACCAAAACAAAACAAAAGCAGCCGAACUGCUCGGCGUUUUAUUUGCACUUCAA (((((((..((((((.(......).))))))..))..............................................(((((.(((((.....))))))))))..)))))...... (-21.26 = -21.70 + 0.44)


| Location | 17,231,041 – 17,231,157 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -22.20 |
| Energy contribution | -21.64 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17231041 116 + 23771897 CAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGCACUUUGCAGUUGGUGCAUAUUGUU--UUUUUUGCGCGACCAGUUUCCCCU--UUUUU ......((((.((((((.((.....)).))))..(((((...................(((((.....)))))....(((.......--.....))))))))..)).))))..--..... ( -29.40) >DroSec_CAF1 115062 118 + 1 CAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGUACUUUGCAGUUGGUGCAUAUUUUUCUUUUUUUUCGCGACCAGUUUCCCCU--UUUUU ......((((.((.((((.((.((.....)).)).))))...................(((((.....)))))((((((..................)).)))))).))))..--..... ( -25.07) >DroSim_CAF1 108856 118 + 1 CAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGCACUUUGCAGUUGGUGCAUAUUGUUUCUUUUUUUCGCGACCAGUUUCCCCU--UUUUU ......((((.((((((.((.....)).))))..(((((...................(((((.....))))).((.((.....)).))........)))))..)).))))..--..... ( -27.50) >DroEre_CAF1 118383 109 + 1 CAUUUCGGGACGUGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGCACUUUGCAGUUUGUGCAUAUU--U----UUUUU--CGACCAGUUUCCCCU---UUUU ......((((((..(((.((.....)).)))..))))(((((................(((((.....))))).))))).....--.----.....--...))..........---.... ( -25.73) >DroYak_CAF1 121190 114 + 1 CAUUUCGGGACAUGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGCACUUUGCAGUUUGUGCAUAUUCUU----UUUUU--CGACCAGUUUCCCCUCUUUUUU ......((((..(((((((((......(((......)))........((((.......(((((.....)))))))))..........----.))))--)).)))...))))......... ( -25.41) >consensus CAUUUCGGGACACGGCGAGGAUACUCCGCGUUUCGUCGCAUAUUUCAACAAAUUUUUUACUGCACUUUGCAGUUGGUGCAUAUUGUU__UUUUUUUCGCGACCAGUUUCCCCU__UUUUU ......((((.((.((((.((.((.....)).)).))))...................(((((.....)))))...............................)).))))......... (-22.20 = -21.64 + -0.56)


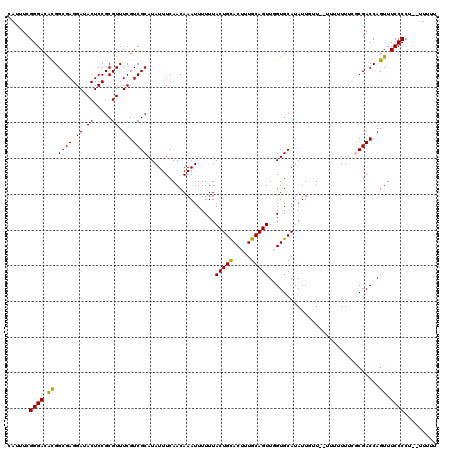
| Location | 17,231,041 – 17,231,157 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -25.28 |
| Energy contribution | -25.24 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.87 |
| SVM RNA-class probability | 0.999672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17231041 116 - 23771897 AAAAA--AGGGGAAACUGGUCGCGCAAAAAA--AACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUG .....--..((((.....((((((.......--(((((.........(((((.....))))).......)))))......))))))....(((((.(......).)))))))))...... ( -30.81) >DroSec_CAF1 115062 118 - 1 AAAAA--AGGGGAAACUGGUCGCGAAAAAAAAGAAAAAUAUGCACCAACUGCAAAGUACAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUG .....--..((((.((.(((((((................((((.....))))...............(((((((.......))))))).))))(((....)))))))).))))...... ( -26.40) >DroSim_CAF1 108856 118 - 1 AAAAA--AGGGGAAACUGGUCGCGAAAAAAAGAAACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUG .....--..((((.((.(((((((.......................(((((.....)))))......(((((((.......))))))).))))(((....)))))))).))))...... ( -30.10) >DroEre_CAF1 118383 109 - 1 AAAA---AGGGGAAACUGGUCG--AAAAA----A--AAUAUGCACAAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCACGUCCCGAAAUG ....---..((((...(((.((--(....----.--.....((....(((((.....)))))......(((((((.......)))))))..))((.....))))))))..))))...... ( -28.50) >DroYak_CAF1 121190 114 - 1 AAAAAAGAGGGGAAACUGGUCG--AAAAA----AAGAAUAUGCACAAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCAUGUCCCGAAAUG .........((((...(((.((--(....----........((....(((((.....)))))......(((((((.......)))))))..))((.....))))))))..))))...... ( -28.50) >consensus AAAAA__AGGGGAAACUGGUCGCGAAAAAAA__AACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUAUGCGACGAAACGCGGAGUAUCCUCGCCGUGUCCCGAAAUG .........((((...((((....................(((....(((((.....)))))......(((((((.......)))))))..)))(((....)))))))..))))...... (-25.28 = -25.24 + -0.04)

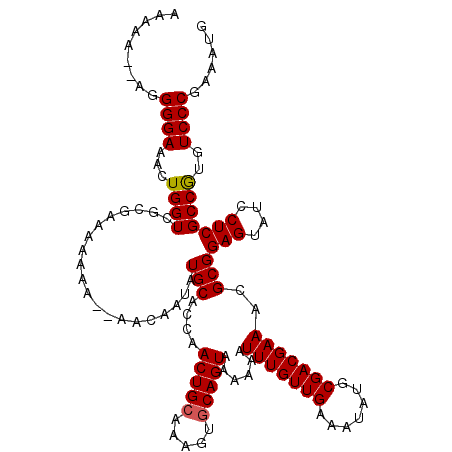
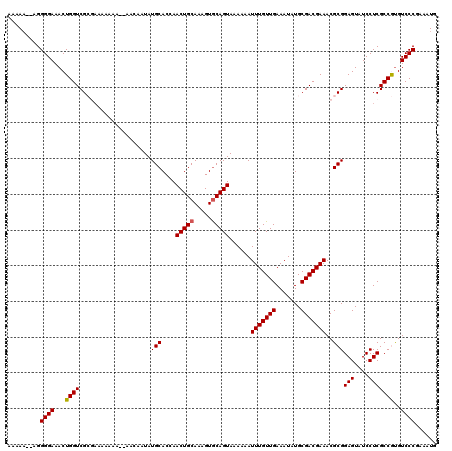
| Location | 17,231,081 – 17,231,197 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.95 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.26 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17231081 116 - 23771897 ACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGCAAAAAA--AACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUA ...(((((.(((((.(((...(....)......(((((.(.....--.)((....)).)))))........--........))))))(((((.....))))).........)).))))). ( -25.90) >DroSec_CAF1 115102 118 - 1 ACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGAAAAAAAAGAAAAAUAUGCACCAACUGCAAAGUACAGUAAAAAAUUUGUUGAAAUA ...(((((.((..(((((...((((.(((....(((((.(.....--.)((....)).)))))................))))))).(((....)))))))).........)).))))). ( -22.55) >DroSim_CAF1 108896 118 - 1 ACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGAAAAAAAGAAACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUA ...(((((.((....(((((((....)......(((((.(.....--.)((....)).)))))........))))))....))....(((((.....)))))............))))). ( -29.00) >DroEre_CAF1 118423 109 - 1 ACCAUUUCCGCUGGCUGUUUCAGUGACAUUUAAGCGACACAAAA---AGGGGAAACUGGUCG--AAAAA----A--AAUAUGCACAAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUA .(((.......))).((((((((((.(((.....((((.(....---.)((....)).))))--.....----.--...))))))..(((((.....)))))...........))))))) ( -20.94) >DroYak_CAF1 121230 114 - 1 ACCAUUUCCGCUGGCUGUUUCAGUGACAUUUAAGCGACACAAAAAAGAGGGGAAACUGGUCG--AAAAA----AAGAAUAUGCACAAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUA ...(((((((((...((((.....))))....))))..(((((......((....)).....--.....----..............(((((.....)))))......))))).))))). ( -20.10) >consensus ACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA__AGGGGAAACUGGUCGCGAAAAAAA__AACAAUAUGCACCAACUGCAAAGUGCAGUAAAAAAUUUGUUGAAAUA .(((.......))).((((((((..(.((((..(((((.(........)((....)).)))))........................(((((.....)))))...)))))..)))))))) (-19.90 = -20.26 + 0.36)

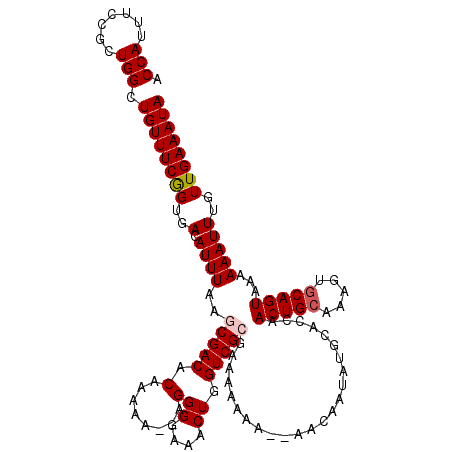

| Location | 17,231,121 – 17,231,232 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -18.58 |
| Energy contribution | -19.58 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17231121 111 - 23771897 CUUAAAUUUUAGCAUAACAAAC-----GAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGCAAAAAA--AACAAUA ......((((.((.........-----(((.....)))(((((((((((.((...(((.(((.(........).))).)))....--)).))))).)))))).)).)))).--....... ( -25.00) >DroSec_CAF1 115142 113 - 1 CUUAAAUUUUAGCAUAACAAAC-----GAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGAAAAAAAAGAAAAAUA ......................-----.......(((((((((((((((.((...(((.(((.(........).))).)))....--)).))))).)))))).))))............. ( -26.30) >DroSim_CAF1 108936 113 - 1 CUUAAAUUUUAGCAUAACAAAC-----GAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA--AGGGGAAACUGGUCGCGAAAAAAAGAAACAAUA ......................-----.......(((((((((((((((.((...(((.(((.(........).))).)))....--)).))))).)))))).))))............. ( -26.30) >DroEre_CAF1 118463 104 - 1 CUUAAAUUUUAGCAUAACAAAC-----GAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCAGUGACAUUUAAGCGACACAAAA---AGGGGAAACUGGUCG--AAAAA----A--AAUA ......................-----...........(((((((((((.((...(((.((.((.........)))).)))...---)).))))).))))))--.....----.--.... ( -20.60) >DroYak_CAF1 121270 114 - 1 CUUAAAUUUUAGCAUAACAAACGAAACGAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCAGUGACAUUUAAGCGACACAAAAAAGAGGGGAAACUGGUCG--AAAAA----AAGAAUA .....(((((............((((........))))((((((((((((((...((((.....))))....)))....(......)...))))).))))))--.....----.))))). ( -18.90) >consensus CUUAAAUUUUAGCAUAACAAAC_____GAAAUACUUUCCGACCAUUUCCGCUGGCUGUUUCGGUGACAUUUAAGCGACACAAAAA__AGGGGAAACUGGUCGCGAAAAAAA__AACAAUA ..................................(((((((((((((((.((...(((.(((.(........).))).)))......)).))))).)))))).))))............. (-18.58 = -19.58 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:32 2006