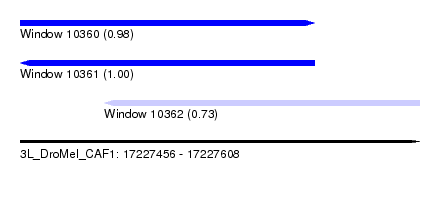
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,227,456 – 17,227,608 |
| Length | 152 |
| Max. P | 0.998945 |

| Location | 17,227,456 – 17,227,568 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -29.10 |
| Energy contribution | -28.94 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976273 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17227456 112 + 23771897 AC--AAGGCCCAAUGCUGGCUGUU------GCUGUUGGCGACAGCACUUGGCUUCGACUACCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ..--..(((.........((((((------((.....))))))))..(((((((..(((((....))))...)..)))))))..)))......(((......)))............... ( -30.40) >DroSec_CAF1 110879 112 + 1 AC--AAGGCCCAAUGCUGGCUGUU------GCUGUUGGCGACAGCACUUGGCUUCGACUACCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ..--..(((.........((((((------((.....))))))))..(((((((..(((((....))))...)..)))))))..)))......(((......)))............... ( -30.40) >DroSim_CAF1 104600 112 + 1 AC--AAGGCCCAAUGCUGGCUGUU------GCUGUUGGCGACAGCACUUGGCUUCGACUACCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ..--..(((.........((((((------((.....))))))))..(((((((..(((((....))))...)..)))))))..)))......(((......)))............... ( -30.40) >DroEre_CAF1 114743 110 + 1 ----AAGGCCUGAUGCUGGCUGUU------GCUGUUGGCGACAGCACUUGGCUUCGACUACCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ----..(((.((((..((((((((------((.....))))))))..(((((((..(((((....))))...)..))))))).......))..)))).)))................... ( -36.30) >DroYak_CAF1 117581 120 + 1 ACAGAAGGCUCGAUGCUGGCUGUUGCUGUUGCUGUUGGCGACAGCACUUGGCUUCGACUGCCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ......(((..(((...(((...(((((((((.....))))))))).(((((((..(((((....))))...)..)))))))..)))......)))..)))................... ( -37.20) >consensus AC__AAGGCCCAAUGCUGGCUGUU______GCUGUUGGCGACAGCACUUGGCUUCGACUACCCUGGUAGCCAUUAAAGCCAAUUGCCCACAAAAUCAUGCCCGAUAUCAUAAUUGCUCUU ......(((....(((..((.((.......)).))..)))...(((.(((((((..(((((....))))...)..))))))).)))............)))................... (-29.10 = -28.94 + -0.16)

| Location | 17,227,456 – 17,227,568 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.97 |
| Mean single sequence MFE | -39.26 |
| Consensus MFE | -35.20 |
| Energy contribution | -35.00 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17227456 112 - 23771897 AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGUCGCCAACAGC------AACAGCCAGCAUUGGGCCUU--GU (((.(((((((((........))))))).(((.(((..((((((((...((((((....)))))))))))))).((((((.....)))))------)...))).)))....)))))--.. ( -38.90) >DroSec_CAF1 110879 112 - 1 AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGUCGCCAACAGC------AACAGCCAGCAUUGGGCCUU--GU (((.(((((((((........))))))).(((.(((..((((((((...((((((....)))))))))))))).((((((.....)))))------)...))).)))....)))))--.. ( -38.90) >DroSim_CAF1 104600 112 - 1 AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGUCGCCAACAGC------AACAGCCAGCAUUGGGCCUU--GU (((.(((((((((........))))))).(((.(((..((((((((...((((((....)))))))))))))).((((((.....)))))------)...))).)))....)))))--.. ( -38.90) >DroEre_CAF1 114743 110 - 1 AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGUCGCCAACAGC------AACAGCCAGCAUCAGGCCUU---- ..................((((.(((...(((.(((..((((((((...((((((....)))))))))))))).((((((.....)))))------)...))).)))))).)))).---- ( -40.20) >DroYak_CAF1 117581 120 - 1 AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGCAGUCGAAGCCAAGUGCUGUCGCCAACAGCAACAGCAACAGCCAGCAUCGAGCCUUCUGU ..................((((.(((...(((.(((..(((((((..((.(((.....))).))..))))))).((((((.((.....)).))))))...))).)))))).))))..... ( -39.40) >consensus AAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGUCGCCAACAGC______AACAGCCAGCAUUGGGCCUU__GU ..................((((.(((...(((.(((..((((((((...((((((....))))))))))))))..(((((.....)))))..........))).)))))).))))..... (-35.20 = -35.00 + -0.20)

| Location | 17,227,488 – 17,227,608 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -35.48 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17227488 120 - 23771897 CUCCCGCACAGCUGCCGGAUUACAGGGUAUGGAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGU ...(((((...(((((.(((..((.(((......))).....((....)).))..))).)))).)....)))))(((.((((((((...((((((....)))))))))))))).)))... ( -39.70) >DroSec_CAF1 110911 120 - 1 CUCCCGCACAGCUGCCGGAUUACAGGGUAUGGAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGU ...(((((...(((((.(((..((.(((......))).....((....)).))..))).)))).)....)))))(((.((((((((...((((((....)))))))))))))).)))... ( -39.70) >DroSim_CAF1 104632 120 - 1 CUCCCGCACAGCUGCCGGAUUACAGGGUAUGGAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGU ...(((((...(((((.(((..((.(((......))).....((....)).))..))).)))).)....)))))(((.((((((((...((((((....)))))))))))))).)))... ( -39.70) >DroEre_CAF1 114773 120 - 1 CUCGCGGACAGCUGCCGGAUUACAGGGUAUGGAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGU ..((((((...(((((.(((..((.(((......))).....((....)).))..))).)))).)..))))))((((.((((((((...((((((....)))))))))))))).)))).. ( -39.20) >DroYak_CAF1 117621 120 - 1 CGCCUGCACAGCUGUUGGAUUACAGGGUAUGCAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGCAGUCGAAGCCAAGUGCUGU .((.(((....((((......)))).))).))...(((((((.....((((((........)))))))))))))(((.(((((((..((.(((.....))).))..))))))).)))... ( -36.70) >consensus CUCCCGCACAGCUGCCGGAUUACAGGGUAUGGAAACCACGAAGAGCAAUUAUGAUAUCGGGCAUGAUUUUGUGGGCAAUUGGCUUUAAUGGCUACCAGGGUAGUCGAAGCCAAGUGCUGU ...(((((...(((((.(((..((.(((......))).....((....)).))..))).)))).)....)))))(((.((((((((...((((((....)))))))))))))).)))... (-35.48 = -35.60 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:16 2006