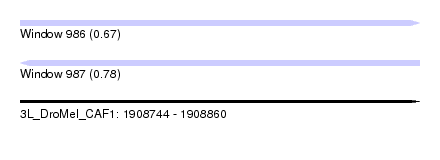
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,908,744 – 1,908,860 |
| Length | 116 |
| Max. P | 0.777449 |

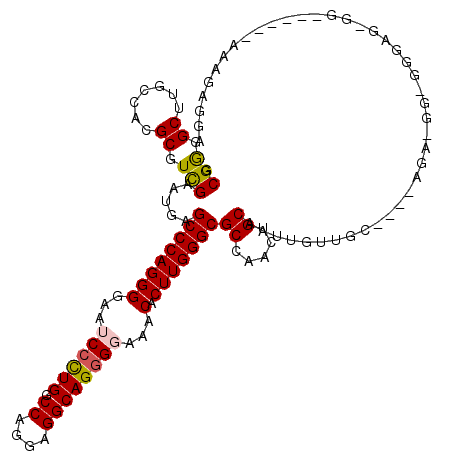
| Location | 1,908,744 – 1,908,860 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -24.13 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1908744 116 + 23771897 UCCUCUUUCCUCUUCCCCUCCCUCCAUCG----GCAACAAUUGCAGUUGGCGCCCAAGUGUUCCCCCUGCCUCCUGGCCAGGGAUUCCCCUGGGCCCAUUCGACGCGUGGCAAGCCUCGC ..........................(((----(((.....)))...(((.(((((.(.(....(((((((....)).)))))...).).))))))))..))).(((.((....)).))) ( -34.10) >DroEre_CAF1 32736 114 + 1 UCCUCAUUU-----CCCGUUCCCCC-UCUGGUUGCAACAAUUGCAGUUGGCGCCCAAGUGUUUCCCCUGCCGCCUGGCCAGGGAUUCCCCUGGGCGCACUCGACGCGUGGCAAGCCCCGC .........-----...(((.....-..(..(((((.....)))))..)(((((((.(.(..((((..(((....)))..))))..).).)))))))....)))(((.((....)).))) ( -39.00) >DroWil_CAF1 46716 98 + 1 -CCAUUUU------ACAAUU---------------UCCAAUUGCAGUGGGCGCCCAAGUGUUUCCCCUGCCACCUGGUCAGGGCUUCCCCUGGGCGCAUUCAACGCGUGGCAAGCCUCGU -(((((..------.((((.---------------....)))).)))))(((((((.(.(....(((((((....)).)))))...).).))))))).......(((.((....)).))) ( -35.10) >DroYak_CAF1 31997 109 + 1 UCCUCUUU------CCACUCUCUCC-UCU----GCAACAAUUGCAGUUGGCGCCCAAGUGUUCCCCCUGCCUCCUGGCCAGGGAUUCCCCUGGGCUCACUCGACGCGUGGCAAGCCCCGC ....(((.------((((.....(.-.((----(((.....)))))..)(((..(.((((..(((...(((....))).((((....)))))))..)))).).))))))).)))...... ( -35.10) >DroAna_CAF1 35776 108 + 1 UCCUUU---CUG---C-AACCCCAC-CUU----GCACCCAUUGCAGUUGGCGCCCAAGUGUUCCCACUGCCCCCUGGCCAAGGAUUCCCCUGGGCUCACUCGACGCGUGGCAAGCCCCGA ......---..(---(-....((((-.((----(((.....)))))...(((..(.((((..(((...(((....)))..(((.....))))))..)))).).)))))))...))..... ( -27.60) >DroPer_CAF1 35078 95 + 1 UCCCCC---AU----------------------GUUGCAAUUGCAGUUGGCGCCCAAGUGUUUCCCCUGCCGCCGGGCCAGGGCUUCCCCUGGGCCCAUUCGACGCGUGGCAAGCCCCGA ....((---((----------------------(((((....)))(((((((....((........))..))))(((((((((....)))).)))))....))))))))).......... ( -33.30) >consensus UCCUCUUU______CC_AUCCC_CC_UCU____GCAACAAUUGCAGUUGGCGCCCAAGUGUUCCCCCUGCCGCCUGGCCAGGGAUUCCCCUGGGCCCACUCGACGCGUGGCAAGCCCCGC .............................................(((((.(((((.(.(....(((((((....)).)))))...).).)))))....)))))(((.((....)).))) (-24.13 = -24.88 + 0.75)

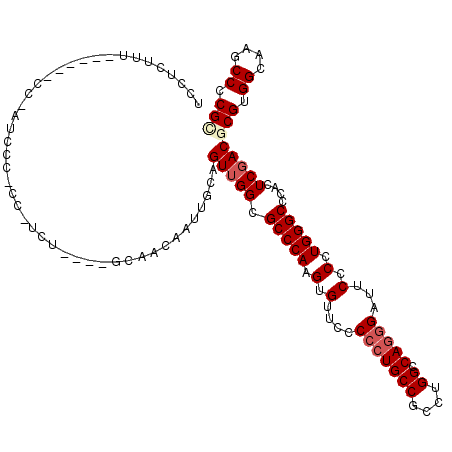
| Location | 1,908,744 – 1,908,860 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -41.68 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 1908744 116 - 23771897 GCGAGGCUUGCCACGCGUCGAAUGGGCCCAGGGGAAUCCCUGGCCAGGAGGCAGGGGGAACACUUGGGCGCCAACUGCAAUUGUUGC----CGAUGGAGGGAGGGGAAGAGGAAAGAGGA ......(((.((.(.(((((..(((((((((((...((((((.((....))))))))...).))))))).)))...(((.....)))----)))))).)).)))................ ( -43.40) >DroEre_CAF1 32736 114 - 1 GCGGGGCUUGCCACGCGUCGAGUGCGCCCAGGGGAAUCCCUGGCCAGGCGGCAGGGGAAACACUUGGGCGCCAACUGCAAUUGUUGCAACCAGA-GGGGGAACGGG-----AAAUGAGGA ((((.(((((.(....).)))))(((((((((....(((((.(((....))).)))))....)))))))))...))))..(((((.(..(....-)..).))))).-----......... ( -44.90) >DroWil_CAF1 46716 98 - 1 ACGAGGCUUGCCACGCGUUGAAUGCGCCCAGGGGAAGCCCUGACCAGGUGGCAGGGGAAACACUUGGGCGCCCACUGCAAUUGGA---------------AAUUGU------AAAAUGG- .((((.((((((((((((.....)))).(((((....))))).....))))))))(....).)))).....(((.((((((....---------------.)))))------)...)))- ( -37.70) >DroYak_CAF1 31997 109 - 1 GCGGGGCUUGCCACGCGUCGAGUGAGCCCAGGGGAAUCCCUGGCCAGGAGGCAGGGGGAACACUUGGGCGCCAACUGCAAUUGUUGC----AGA-GGAGAGAGUGG------AAAGAGGA ......(((.((((((((((((((..(((((((....)))).(((....)))...)))..))))).)))))...(((((.....)))----)).-.......))))------...))).. ( -43.50) >DroAna_CAF1 35776 108 - 1 UCGGGGCUUGCCACGCGUCGAGUGAGCCCAGGGGAAUCCUUGGCCAGGGGGCAGUGGGAACACUUGGGCGCCAACUGCAAUGGGUGC----AAG-GUGGGGUU-G---CAG---AAAGGA ((...((..(((.(((.(((((((..(((((((....)))).(((....)))...)))..))))))))))(((..((((.....)))----)..-.)))))).-)---).)---)..... ( -38.60) >DroPer_CAF1 35078 95 - 1 UCGGGGCUUGCCACGCGUCGAAUGGGCCCAGGGGAAGCCCUGGCCCGGCGGCAGGGGAAACACUUGGGCGCCAACUGCAAUUGCAAC----------------------AU---GGGGGA (((((((((.((..(.(((.....))).)..)).)))))))))(((((((.(((((....).))).).))))...(((....)))..----------------------..---)))... ( -42.00) >consensus GCGGGGCUUGCCACGCGUCGAAUGAGCCCAGGGGAAUCCCUGGCCAGGAGGCAGGGGAAACACUUGGGCGCCAACUGCAAUUGUUGC____AGA_GG_GGGAG_GG______AAAGAGGA .(((.((.......)).))).....((((((((...((((((.((....))))))))...).)))))))((.....)).......................................... (-29.90 = -29.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:47:30 2006