
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,226,439 – 17,226,704 |
| Length | 265 |
| Max. P | 0.999331 |

| Location | 17,226,439 – 17,226,544 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -25.37 |
| Consensus MFE | -21.02 |
| Energy contribution | -21.42 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226439 105 + 23771897 UGUGUCUGUGUGUGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCCA-------ACC-------- (((((.((((.(((.....))).))))..)))))..................(((((.....(((......)))..((((((((....))))))))..))))-------)..-------- ( -31.90) >DroSec_CAF1 109863 92 + 1 ------------UGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGUACUCGGAUCCGCCCCC--------ACC-------- ------------((((..............))))...................((((.....(((......)))..((((((((....))))))))..)))--------)..-------- ( -24.64) >DroSim_CAF1 103566 92 + 1 ------------UGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCC--------ACC-------- ------------((((..............))))...................((((.....(((......)))..((((((((....))))))))..)))--------)..-------- ( -24.34) >DroEre_CAF1 113789 100 + 1 ------------UGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGAAUCCGCCCACACCACUCCACC-------- ------------((((..............))))...................(((((.((((((......)))((((....))))...)))...)))))............-------- ( -19.74) >DroYak_CAF1 116530 108 + 1 ------------UGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGGCUCGUAUCCGCCCACACCGAUCCACCGACCCACU ------------((((..............))))...................((((((...(((......)))...(((((.((((........)))).....)))))...)))))).. ( -26.24) >consensus ____________UGUGCCACGCUUAUAAAACACAUUUAUCAUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCCA_______ACC________ ............((((..............))))....................((((....(((......)))...(((((((....)))))))))))..................... (-21.02 = -21.42 + 0.40)

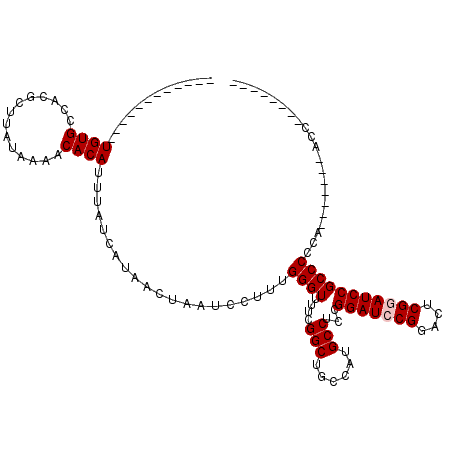
| Location | 17,226,439 – 17,226,544 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -26.82 |
| Energy contribution | -26.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226439 105 - 23771897 --------GGU-------UGGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACACACACAGACACA --------..(-------((((..((((((((....))))))))..(((......))).....)))))..................(((((((.....(.(((.....))).)))))))) ( -34.00) >DroSec_CAF1 109863 92 - 1 --------GGU--------GGGGGCGGAUCCGAGUACGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACA------------ --------..(--------(((..((((((((....))))))))..(((......))).....))))........(((((((((((.....)))))...))))))...------------ ( -30.10) >DroSim_CAF1 103566 92 - 1 --------GGU--------GGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACA------------ --------..(--------(((..((((((((....))))))))..(((......))).....))))........(((((((((((.....)))))...))))))...------------ ( -30.50) >DroEre_CAF1 113789 100 - 1 --------GGUGGAGUGGUGUGGGCGGAUUCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACA------------ --------.........(((((((((((((((....))))))))..(((......))).....))).........(((...(((((.....))))).)))...)))).------------ ( -27.30) >DroYak_CAF1 116530 108 - 1 AGUGGGUCGGUGGAUCGGUGUGGGCGGAUACGAGCCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACA------------ ..(((((...((((((....(((((........)))))))))))..(((......)))....)))))........(((((((((((.....)))))...))))))...------------ ( -31.10) >consensus ________GGU_______UGGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAUGAUAAAUGUGUUUUAUAAGCGUGGCACA____________ .....................(((((((((((....))))))))..(((......))).....))).........(((((((((((.....)))))...))))))............... (-26.82 = -26.90 + 0.08)



| Location | 17,226,479 – 17,226,584 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -22.44 |
| Energy contribution | -22.84 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226479 105 + 23771897 AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCCA-------ACC--------GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA ............(((((.....(((......)))..((((((((....))))))))..))))-------).(--------((....((......))..)))................... ( -26.80) >DroSec_CAF1 109891 104 + 1 AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGUACUCGGAUCCGCCCCC--------ACC--------GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA .............((((.....(((......)))..((((((((....))))))))..)))--------).(--------((....((......))..)))................... ( -26.20) >DroSim_CAF1 103594 104 + 1 AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCC--------ACC--------GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA .............((((.....(((......)))..((((((((....))))))))..)))--------).(--------((....((......))..)))................... ( -25.90) >DroEre_CAF1 113817 112 + 1 AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGAAUCCGCCCACACCACUCCACC--------GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA .............(((((.((((((......)))((((....))))...)))...)))))...........(--------((....((......))..)))................... ( -21.30) >DroYak_CAF1 116558 120 + 1 AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGGCUCGUAUCCGCCCACACCGAUCCACCGACCCACUGCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA .......(((...((((((...(((......)))...(((((.((((........)))).....)))))...))))))...........(((.....))))))................. ( -27.70) >consensus AUAACUAAUCCUUUGGGUUUUCGGCUGCCAUGCCUCCGGAUCCGGACUCGGAUCCGCCCCCA_______ACC________GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAA .......(((....((((....(((......)))...(((((((....)))))))))))..............................(((.....))))))................. (-22.44 = -22.84 + 0.40)



| Location | 17,226,479 – 17,226,584 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -34.76 |
| Energy contribution | -34.84 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.52 |
| SVM RNA-class probability | 0.999331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226479 105 - 23771897 UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGC--------GGU-------UGGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU ..((((.((....(((((((((((......))))))))))--------).(-------((((..((((((((....))))))))..(((......))).....)))))..)).))))... ( -40.90) >DroSec_CAF1 109891 104 - 1 UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGC--------GGU--------GGGGGCGGAUCCGAGUACGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU ..((((.((....(((((((((((......))))))))))--------).(--------(((..((((((((....))))))))..(((......))).....))))...)).))))... ( -39.60) >DroSim_CAF1 103594 104 - 1 UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGC--------GGU--------GGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU ..((((.((....(((((((((((......))))))))))--------).(--------(((..((((((((....))))))))..(((......))).....))))...)).))))... ( -40.00) >DroEre_CAF1 113817 112 - 1 UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGC--------GGUGGAGUGGUGUGGGCGGAUUCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU .((((((((..(.(((((((((((......))))))))))--------).).))))))))((((((((((((....))))))))..(((......))).....))))............. ( -40.80) >DroYak_CAF1 116558 120 - 1 UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGCAGUGGGUCGGUGGAUCGGUGUGGGCGGAUACGAGCCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU ..((((.((....(((((((((((......))))))))))).(((((...((((((....(((((........)))))))))))..(((......)))....)))))...)).))))... ( -41.30) >consensus UUAUUAUUCAAAGUGUUAUCGCUGACAGAGCAGUGGUGGC________GGU_______UGGGGGCGGAUCCGAGUCCGGAUCCGGAGGCAUGGCAGCCGAAAACCCAAAGGAUUAGUUAU ..((((.((.....((((((((((......)))))))))).....................(((((((((((....))))))))..(((......))).....)))....)).))))... (-34.76 = -34.84 + 0.08)

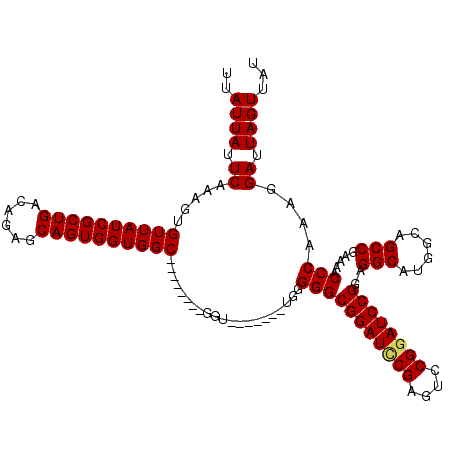

| Location | 17,226,544 – 17,226,664 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -24.51 |
| Energy contribution | -25.71 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226544 120 + 23771897 GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAA ((((....((((.....))))......(((((...................)))))......))))...(((((....)))))...(((((.(......).))))).............. ( -30.71) >DroSec_CAF1 109955 120 + 1 GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAA ((((....((((.....))))......(((((...................)))))......))))...(((((....)))))...(((((.(......).))))).............. ( -30.71) >DroSim_CAF1 103658 120 + 1 GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAA ((((....((((.....))))......(((((...................)))))......))))...(((((....)))))...(((((.(......).))))).............. ( -30.71) >DroEre_CAF1 113889 116 + 1 GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGG----CAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCAAA ((((....((((.....))))......(((((...................)))))......))))...((----(....)))...(((((.(......).))))).............. ( -23.41) >DroYak_CAF1 116638 116 + 1 GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGG----CAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCAAA ((((....((((.....))))......(((((...................)))))......))))...((----(....)))...(((((.(......).))))).............. ( -23.41) >consensus GCCACCACUGCUCUGUCAGCGAUAACACUUUGAAUAAUAAAUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAA ((((....((((.....))))......(((((...................)))))......))))...(((((....)))))...(((((.(......).))))).............. (-24.51 = -25.71 + 1.20)

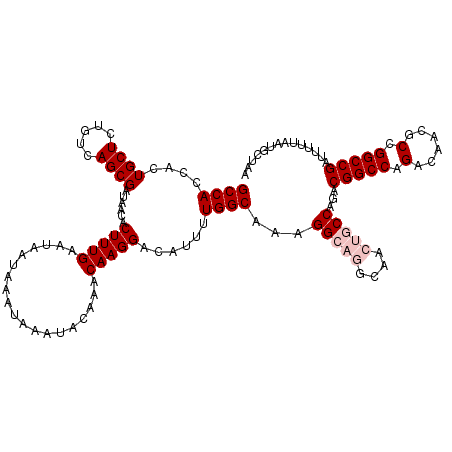

| Location | 17,226,584 – 17,226,704 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -25.06 |
| Energy contribution | -26.66 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226584 120 + 23771897 AUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............((.(...((((((..(((((....)))))...(((((.(......).)))))........))))))...............(((((.((....)).))))).))). ( -32.90) >DroSec_CAF1 109995 120 + 1 AUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............((.(...((((((..(((((....)))))...(((((.(......).)))))........))))))...............(((((.((....)).))))).))). ( -32.90) >DroSim_CAF1 103698 120 + 1 AUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............((.(...((((((..(((((....)))))...(((((.(......).)))))........))))))...............(((((.((....)).))))).))). ( -32.90) >DroEre_CAF1 113929 116 + 1 AUAAAUACAAACAAGGACAUUUUGGCAAAGG----CAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCAAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............(((((((((((((..(.----...))))))))(((((.(......).)))))......................))))...(((((.((....)).)))))..)). ( -25.20) >DroYak_CAF1 116678 116 + 1 AUAAAUACAAACAAGGACAUUUUGGCAAAGG----CAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCAAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............(((((((((((((..(.----...))))))))(((((.(......).)))))......................))))...(((((.((....)).)))))..)). ( -25.20) >consensus AUAAAUACAAACAAGGACAUUUUGGCAAAGGCAGGCAACUGCCAGACGGCCAGACAACGCCGGCCGAUUUUUAAUGCUAACGAUUAUCAUGUAAGGCAAUAUAAAAAUAAAUUGCAGCCG ..............((.(...((((((..(((((....)))))...(((((.(......).)))))........))))))...............(((((.((....)).))))).))). (-25.06 = -26.66 + 1.60)



| Location | 17,226,584 – 17,226,704 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.49 |
| Mean single sequence MFE | -28.90 |
| Consensus MFE | -26.76 |
| Energy contribution | -26.76 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17226584 120 - 23771897 CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUAGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUGCCUGCCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU .(((((......(((((((...(((...((((....)).))..))).))))))))))))((((((...((((.....(((((....)))))...)))).))))))............... ( -29.50) >DroSec_CAF1 109995 120 - 1 CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUAGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUGCCUGCCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU .(((((......(((((((...(((...((((....)).))..))).))))))))))))((((((...((((.....(((((....)))))...)))).))))))............... ( -29.50) >DroSim_CAF1 103698 120 - 1 CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUAGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUGCCUGCCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU .(((((......(((((((...(((...((((....)).))..))).))))))))))))((((((...((((.....(((((....)))))...)))).))))))............... ( -29.50) >DroEre_CAF1 113929 116 - 1 CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUUGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUG----CCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU .....(((((.((....)).)))))..((((.(((((.(((((...........(((((((......)))))))..((((((...----...)))))))))))...)))))))))..... ( -28.00) >DroYak_CAF1 116678 116 - 1 CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUUGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUG----CCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU .....(((((.((....)).)))))..((((.(((((.(((((...........(((((((......)))))))..((((((...----...)))))))))))...)))))))))..... ( -28.00) >consensus CGGCUGCAAUUUAUUUUUAUAUUGCCUUACAUGAUAAUCGUUAGCAUUAAAAAUCGGCCGGCGUUGUCUGGCCGUCUGGCAGUUGCCUGCCUUUGCCAAAAUGUCCUUGUUUGUAUUUAU ((((((((((.((....)).))))).......(((((.(((..((...........))..)))))))).)))))..((((((..(....)..))))))...................... (-26.76 = -26.76 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:12 2006