
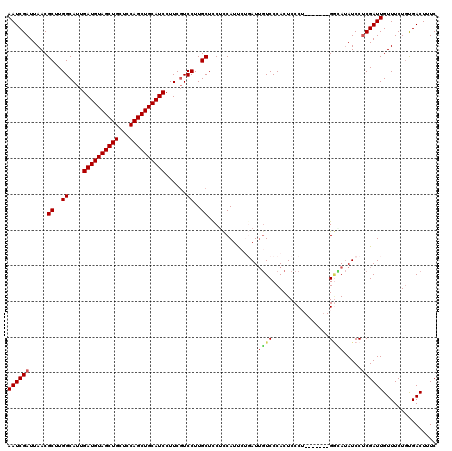
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,225,403 – 17,225,548 |
| Length | 145 |
| Max. P | 0.999067 |

| Location | 17,225,403 – 17,225,516 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.52 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.77 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17225403 113 - 23771897 AAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCUUCCUUGCUUCUCCAUCCUGAUUGUCCCACUCCCU-------GGAAUAUCCUCGAUUGUUUCUGUGACUUUC ((((((.....((..((....((((((((((...))))))))))......))..))...........(((..(((........-------)))..))).))))))............... ( -25.90) >DroSec_CAF1 108830 113 - 1 AAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUCCCU-------GGCAUAUCCUCGAUUGUUUCUGUGACUUUC ((((((.....((..((((..((((((((((...))))))))))..).).))..))...........(((((((.........-------)))).))).))))))............... ( -25.50) >DroSim_CAF1 102571 113 - 1 AAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUCCCU-------GGCAUAUCCUCGAUUGUUUCUGCGACUUUC ((((((.....((..((((..((((((((((...))))))))))..).).))..))...........(((((((.........-------)))).))).))))))............... ( -25.50) >DroEre_CAF1 112616 116 - 1 AAUCGAUUAACGC-UGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCAUCAUU---AUUGUCCCACUCCCACUCCGAGGAUAUAUCCCCGAUUGUUUCUGUGACUUUC ....(((....((-.((((..((((((((((...))))))))))..).)))...))...)))...---...(((.((....((.((.(.(((....)))).)).))....)).))).... ( -24.60) >DroYak_CAF1 115346 110 - 1 AAUCGAUUAACGCCUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUUCAUU---AUGCUCCCACUCCCA-------GCGAAAUCCUCGAUUGUUUCUAUGACUUUC ((((((....(((.(((....((((((((((...))))))))))....((....((.........---..))....))..)))-------)))......))))))............... ( -24.90) >consensus AAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUCCCU_______GGCAUAUCCUCGAUUGUUUCUGUGACUUUC ((((((.....((..((....((((((((((...))))))))))......))..))..............((((................)))).....))))))............... (-19.41 = -19.77 + 0.36)


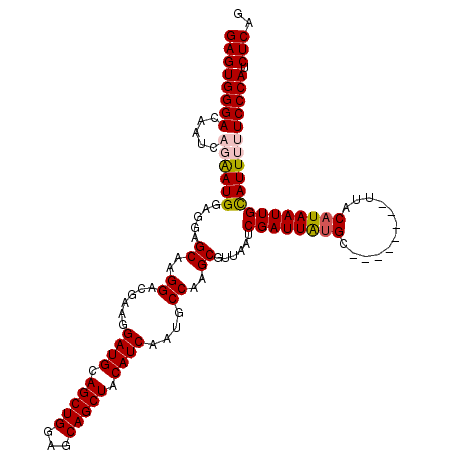
| Location | 17,225,436 – 17,225,548 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -29.54 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17225436 112 + 23771897 GAGUGGGACAAUCAGGAUGGAGAAGCAAGGAAGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCAAGCGUUAAUCGAUUAUGC--------UUACAUAAUUGCAUUUUUCCCAUCUCAG ((((((((.....((((((.....((..((......((((.(((((...))))).))))....))..))......((((((((.--------...))))))))))))))))))).))).. ( -32.90) >DroSec_CAF1 108863 112 + 1 GAGUGGGACAAUCAGAAUGGAGGAGCAAGGACGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCAAGCGUUAAUCGAUUAUGC--------UUACAUAAUUGUAUUUUUCCCAUCUCAA ((((((((.((.(((.(((...(((((..(.(((..((((.(((((...))))).))))(((((...)))))..))).)..)))--------)).)))..)))...)).))))).))).. ( -33.40) >DroSim_CAF1 102604 112 + 1 GAGUGGGACAAUCAGAAUGGAGGAGCAAGGACGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCAAGCGUUAAUCGAUUAUGC--------UUACAUAAUUGCAUUUUUCCCAUCUCAG ((((((((.....((((((.....((..((.((...((((.(((((...))))).))))..))))..))......((((((((.--------...))))))))))))))))))).))).. ( -33.70) >DroEre_CAF1 112656 116 + 1 GAGUGGGACAAU---AAUGAUGGAGCAAGGACGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCA-GCGUUAAUCGAUUGCGCUUGCCGACUUACAUAAUUGCAUUUUUCCCAUCUCGG ((((((((((..---..)).....((((.....(((((((.(((((...))))).))))...((.(-((((.........))))).))...)))......)))).....))))).))).. ( -34.00) >DroYak_CAF1 115379 117 + 1 GAGUGGGAGCAU---AAUGAAGGAGCAAGGACGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCAGGCGUUAAUCGAUUGUGCUUGCCGACUUACAUAAUUGCAUUUUUCCCAUCUCAG (((((((((((.---.((((((..(((((.((((..((((.(((((...))))).))))(((((...)))))......)))).)))))...))).)))...))).....))))).))).. ( -39.80) >consensus GAGUGGGACAAUCAGAAUGGAGGAGCAAGGACGAAGGAUGCAGCUGGAGCAGCUACAUCAAUGCCAAGCGUUAAUCGAUUAUGC________UUACAUAAUUGCAUUUUUCCCAUCUCAG ((((((((.....((((((.....((..((......((((.(((((...))))).))))....))..))......((((((((............))))))))))))))))))).))).. (-29.54 = -29.98 + 0.44)


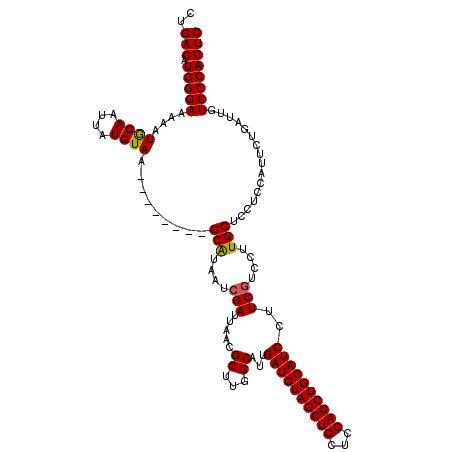
| Location | 17,225,436 – 17,225,548 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.34 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.04 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17225436 112 - 23771897 CUGAGAUGGGAAAAAUGCAAUUAUGUAA--------GCAUAAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCUUCCUUGCUUCUCCAUCCUGAUUGUCCCACUC ..(((.(((((......(.((((((...--------.)))))).)......((..((....((((((((((...))))))))))......))..))................)))))))) ( -31.40) >DroSec_CAF1 108863 112 - 1 UUGAGAUGGGAAAAAUACAAUUAUGUAA--------GCAUAAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUC ..(((.(((((......(.((((((...--------.)))))).)......((..((((..((((((((((...))))))))))..).).))..))................)))))))) ( -32.10) >DroSim_CAF1 102604 112 - 1 CUGAGAUGGGAAAAAUGCAAUUAUGUAA--------GCAUAAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUC ..(((.(((((......(.((((((...--------.)))))).)......((..((((..((((((((((...))))))))))..).).))..))................)))))))) ( -31.80) >DroEre_CAF1 112656 116 - 1 CCGAGAUGGGAAAAAUGCAAUUAUGUAAGUCGGCAAGCGCAAUCGAUUAACGC-UGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCAUCAUU---AUUGUCCCACUC ..(((.(((((..((((.......(((((.((((.((((...........)))-).))...((((((((((...))))))))))...))..))))).....))))---....)))))))) ( -36.40) >DroYak_CAF1 115379 117 - 1 CUGAGAUGGGAAAAAUGCAAUUAUGUAAGUCGGCAAGCACAAUCGAUUAACGCCUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUUCAUU---AUGCUCCCACUC ..(((.(((((.....(((...(((.(((..((((((.((...........((...))...((((((((((...))))))))))....)).)))))).)))))).---.))))))))))) ( -36.80) >consensus CUGAGAUGGGAAAAAUGCAAUUAUGUAA________GCAUAAUCGAUUAACGCUUGGCAUUGAUGUAGCUGCUCCAGCUGCAUCCUUCGUCCUUGCUCCUCCAUUCUGAUUGUCCCACUC ..(((.(((((....((((....)))).........(((....(((.....((...))...((((((((((...))))))))))..)))....)))................)))))))) (-29.16 = -29.04 + -0.12)

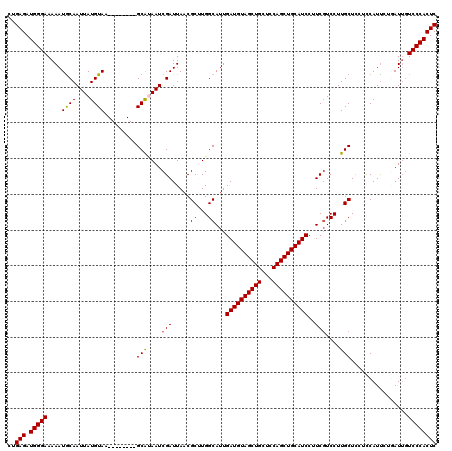
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:05 2006