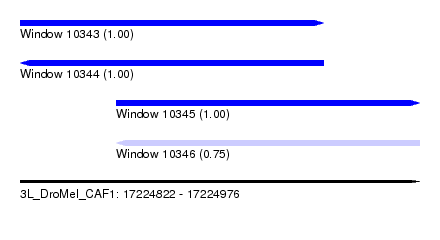
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,224,822 – 17,224,976 |
| Length | 154 |
| Max. P | 0.999891 |

| Location | 17,224,822 – 17,224,939 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.86 |
| Consensus MFE | -38.24 |
| Energy contribution | -38.28 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.38 |
| SVM RNA-class probability | 0.999885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224822 117 + 23771897 CAUUGCUGUUCGGCGGCAGUCAUCGUC---AGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...((((((.(((((((.(((.(((((---((.((.......))....(((........)))........)))))...(((((((......))))))).))))))))))))))))))... ( -43.80) >DroSec_CAF1 108265 120 + 1 CAUUGCCGUUCGGCGGCAGUCAUCGUCACCAGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...(((.((.(((((((.(((...((((...(.((.......)))...)))).....(((((((((.....((((((.....))))))....))))))))))))))))))))).)))... ( -38.20) >DroSim_CAF1 101986 120 + 1 CAUUGCCGUUCGGCGGCAGUCAUCGUCACCAGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...(((.((.(((((((.(((...((((...(.((.......)))...)))).....(((((((((.....((((((.....))))))....))))))))))))))))))))).)))... ( -38.20) >DroEre_CAF1 112008 120 + 1 CAUUGCCGCUCGGCGGCAGUCAUCGUCACCAGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...(((.((.(((((((.(((...((((...(.((.......)))...)))).....(((((((((.....((((((.....))))))....))))))))))))))))))))).)))... ( -40.90) >DroYak_CAF1 114736 120 + 1 CAUUGCCGUUCGGCGGCAGUCAUCGUCACCAGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...(((.((.(((((((.(((...((((...(.((.......)))...)))).....(((((((((.....((((((.....))))))....))))))))))))))))))))).)))... ( -38.20) >consensus CAUUGCCGUUCGGCGGCAGUCAUCGUCACCAGAGCUGCACAUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUU ...(((.((.(((((((.(((...((((...(.((.......)))...)))).....(((((((((.....((((((.....))))))....))))))))))))))))))))).)))... (-38.24 = -38.28 + 0.04)

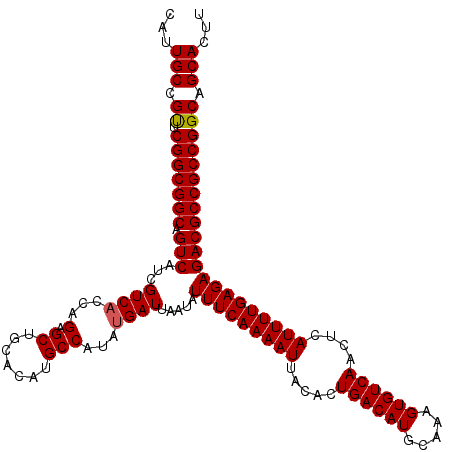

| Location | 17,224,822 – 17,224,939 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -45.48 |
| Consensus MFE | -43.88 |
| Energy contribution | -43.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224822 117 - 23771897 AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCU---GACGAUGACUGCCGCCGAACAGCAAUG ...(((((.(((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).))))))))))---)).)).))).)))))))..)))))... ( -43.00) >DroSec_CAF1 108265 120 - 1 AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCUGGUGACGAUGACUGCCGCCGAACGGCAAUG ...(((((.(((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).)))))))))...))).)).))).)))))))..)))))... ( -44.80) >DroSim_CAF1 101986 120 - 1 AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCUGGUGACGAUGACUGCCGCCGAACGGCAAUG ...(((((.(((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).)))))))))...))).)).))).)))))))..)))))... ( -44.80) >DroEre_CAF1 112008 120 - 1 AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCUGGUGACGAUGACUGCCGCCGAGCGGCAAUG ...(((((((((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).)))))))))...))).)).))).))))))).))))))... ( -50.00) >DroYak_CAF1 114736 120 - 1 AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCUGGUGACGAUGACUGCCGCCGAACGGCAAUG ...(((((.(((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).)))))))))...))).)).))).)))))))..)))))... ( -44.80) >consensus AAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAUGUGCAGCUCUGGUGACGAUGACUGCCGCCGAACGGCAAUG ...(((((.(((((((((((((((....((((((.(((.((((((....))))))...((..((((.....))))..)).)))))))))...))).)).))).)))))))..)))))... (-43.88 = -43.92 + 0.04)


| Location | 17,224,859 – 17,224,976 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -36.06 |
| Consensus MFE | -32.78 |
| Energy contribution | -33.18 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224859 117 + 23771897 AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCAAG---CAGCCUGGCACUUCGGCAC .((((............(((((((((.....((((((.....))))))....)))))))))......((((((....(((((..(....)...)))))---..))).))).....)))). ( -32.80) >DroSec_CAF1 108305 120 + 1 AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCAGGCGGCAGCCUGGCACUUCGGCAC .((((............(((((((((.....((((((.....))))))....)))))))))...((((((((..((.....))...)))))....((((....))))))).....)))). ( -39.20) >DroSim_CAF1 102026 120 + 1 AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCAGGCGGCAGCCUGGCACUUCGGCAC .((((............(((((((((.....((((((.....))))))....)))))))))...((((((((..((.....))...)))))....((((....))))))).....)))). ( -39.20) >DroEre_CAF1 112048 117 + 1 AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCUGG---CAGCCUGGCACUUCGGCAC .((((............(((((((((.....((((((.....))))))....)))))))))......((((((.((.(..((..(....)..))..))---).))).))).....)))). ( -33.30) >DroYak_CAF1 114776 117 + 1 AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCAGG---CACCCUGGCACUUUGGCAC .(((((...........(((((((((.....((((((.....))))))....)))))))))...((((((((......)))))..(((((...)))))---......)))....))))). ( -35.80) >consensus AUGCCAUAUGAUUAAUAUUUCAAAAUUACACUGACAUGCAAAGUGUCAACUCAUUUUGAGAGACGCCGCCGGCAGCACUUGGCACCCUGGCACCCAGG___CAGCCUGGCACUUCGGCAC .((((............(((((((((.....((((((.....))))))....)))))))))...((((((((......)))))..(((((...))))).........))).....)))). (-32.78 = -33.18 + 0.40)

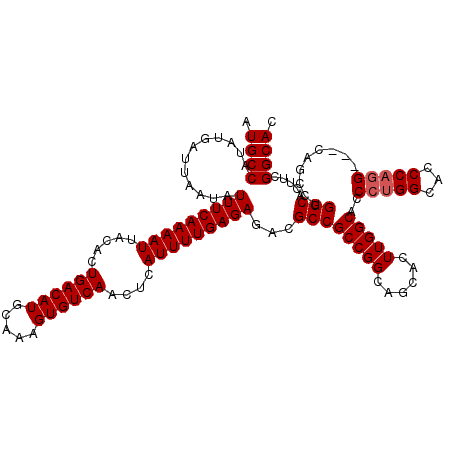
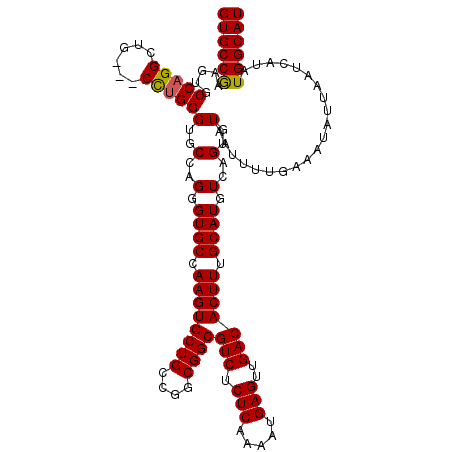
| Location | 17,224,859 – 17,224,976 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.15 |
| Mean single sequence MFE | -40.34 |
| Consensus MFE | -35.66 |
| Energy contribution | -35.54 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224859 117 - 23771897 GUGCCGAAGUGCCAGGCUG---CUUGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU (((((((..((((.(((.(---(...((..(....)..))....)).)))...))))..))(((((((..(((((((.......)))))))...)))))))..............))))) ( -40.80) >DroSec_CAF1 108305 120 - 1 GUGCCGAAGUGCCAGGCUGCCGCCUGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU (((((((..((((.(.(.((.((((.((..(....)..)).)).)).)).).)))))..))(((((((..(((((((.......)))))))...)))))))..............))))) ( -40.80) >DroSim_CAF1 102026 120 - 1 GUGCCGAAGUGCCAGGCUGCCGCCUGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU (((((((..((((.(.(.((.((((.((..(....)..)).)).)).)).).)))))..))(((((((..(((((((.......)))))))...)))))))..............))))) ( -40.80) >DroEre_CAF1 112048 117 - 1 GUGCCGAAGUGCCAGGCUG---CCAGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU (((((((..((((.(((.(---((..((..(....)..))..).)).)))...))))..))(((((((..(((((((.......)))))))...)))))))..............))))) ( -40.90) >DroYak_CAF1 114776 117 - 1 GUGCCAAAGUGCCAGGGUG---CCUGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU ((((((..(..((..((..---(....)..))..))..)((((.((((....))))(((.(((.....)))..)))...((((((....)))))).))))..............)))))) ( -38.40) >consensus GUGCCGAAGUGCCAGGCUG___CCUGGGUGCCAGGGUGCCAAGUGCUGCCGGCGGCGUCUCUCAAAAUGAGUUGACACUUUGCAUGUCAGUGUAAUUUUGAAAUAUUAAUCAUAUGGCAU ((((((.....(((((......)))))(..(..(.((((.((((((((....))))(((.(((.....)))..))))))).)))).)..)..).....................)))))) (-35.66 = -35.54 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:19:01 2006