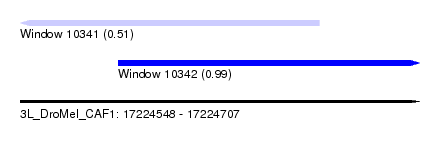
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,224,548 – 17,224,707 |
| Length | 159 |
| Max. P | 0.985034 |

| Location | 17,224,548 – 17,224,667 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.90 |
| Mean single sequence MFE | -29.22 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.506390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224548 119 - 23771897 UAUUACGUUACGGCACACACCAACUCAUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAAAAUGUUCGAGCCAAGCGCACUGGGAGAAAUCAGGU-CAA ...........((......))......(((((...((((..((........))...((.((((((((((..((.....))..))))...)))).)).)).))))((....)).)))-)). ( -28.00) >DroSec_CAF1 108015 120 - 1 UAUUACGUUACGGCACACACUAACUCAUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAAAAUGUUCGAGCCAAGCGCACUGGGAGAAACCAGUUGCAA ............((.........(((.(((.....)))...((........)).)))...(((((((((..((.....))..))))...))))).))(((((((......)))).))).. ( -26.10) >DroSim_CAF1 101736 120 - 1 UAUUACGUUACGGCACACACCAACUCAUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAAAAUGUUCGAGCCAAGUGCACUGGGAGAAAUCAGGUGCAA ............((((...(((.....)))..(..((((..((........))...(((((((((((((..((.....))..))))...)))).))))).))))..).......)))).. ( -31.70) >DroEre_CAF1 111763 113 - 1 UAUUACGUUACGGCACACACCAACUCAUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAA-ACGUUCGAGCCAAGUGCACUGGGAGAAAUC------AA ...........(((.((..........)))))...((((..((........))...(((((((((................-.......)))).))))).))))((....))------.. ( -23.80) >DroYak_CAF1 114484 119 - 1 UAUUACGUUACGGCACACACCGACUGGUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAA-AUGUUCGAGCCAAGUGCACUGGGCGAAAGUAUGGGUAA .................((((....))))(((((.((((..((........))...(((((((((((((..((.....)).-))))...)))).))))).))))((....)).))))).. ( -36.50) >consensus UAUUACGUUACGGCACACACCAACUCAUGGCCCGACCAGAACCAUAACAAAGGAGAGCACUGGCUGCAUAAUCAAAUAGAAAAUGUUCGAGCCAAGUGCACUGGGAGAAAUCAGGUGCAA ...........(((.((..........)))))...((((..((........))...(((((((((((((..((.....))..))))...)))).))))).))))((....))........ (-23.56 = -23.96 + 0.40)

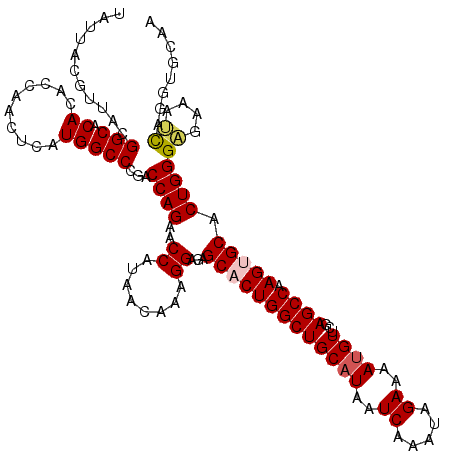
| Location | 17,224,587 – 17,224,707 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.00 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -34.10 |
| Energy contribution | -34.90 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224587 120 + 23771897 UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCAUGAGUUGGUGUGUGCCGUAACGUAAUAUGGAGCAGAAACAAUAGUUUCGGCCUCAUCAUCAUCCAAC ......((((.(((..(((..((((((....(((((((((......((.((((.....)))).)).)))))))))......))))))(((((....))))))))..))).))))...... ( -38.60) >DroSec_CAF1 108055 120 + 1 UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCAUGAGUUAGUGUGUGCCGUAACGUAAUAUGGAGAAGAAACAAUAGUUUCGGCCUAAUCAUCAUCCAAC ......((((((....(((.....(((((..(((((((((.(((((((.....))).)))).....)))))))))......))))).(((((....)))))))).))))))......... ( -30.80) >DroSim_CAF1 101776 120 + 1 UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCAUGAGUUGGUGUGUGCCGUAACGUAAUAUGGAGCAGAAACAAUAGUUUCGGCCUCAUCAUCAUCCAAC ......((((.(((..(((..((((((....(((((((((......((.((((.....)))).)).)))))))))......))))))(((((....))))))))..))).))))...... ( -38.60) >DroEre_CAF1 111796 120 + 1 UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCAUGAGUUGGUGUGUGCCGUAACGUAAUAUGGAGCAGAAACAAUAGUUUCGGCCUCAUCAUCAUCCAAC ......((((.(((..(((..((((((....(((((((((......((.((((.....)))).)).)))))))))......))))))(((((....))))))))..))).))))...... ( -38.60) >DroYak_CAF1 114523 120 + 1 UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCACCAGUCGGUGUGUGCCGUAACGUAAUAUGGAGCAGAAACAAUAGUUUCGGCCUCAUCAUCAUCCAAC ......((((.(((..(((..((((((....(((((((((.((((..((....))..)))).....)))))))))......))))))(((((....))))))))..))).))))...... ( -39.20) >consensus UCUAUUUGAUUAUGCAGCCAGUGCUCUCCUUUGUUAUGGUUCUGGUCGGGCCAUGAGUUGGUGUGUGCCGUAACGUAAUAUGGAGCAGAAACAAUAGUUUCGGCCUCAUCAUCAUCCAAC ......((((.(((..(((..((((((....(((((((((......((.((((.....)))).)).)))))))))......))))))(((((....))))))))..))).))))...... (-34.10 = -34.90 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:57 2006