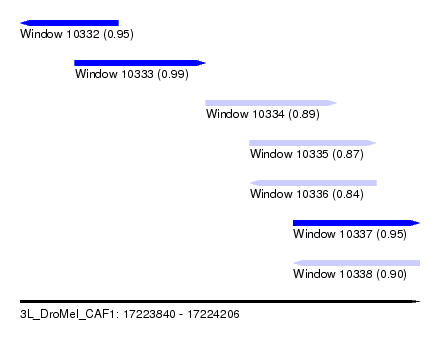
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,223,840 – 17,224,206 |
| Length | 366 |
| Max. P | 0.991093 |

| Location | 17,223,840 – 17,223,930 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 84.26 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -26.17 |
| Energy contribution | -28.67 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17223840 90 - 23771897 GAUGAGAGUGAAUUGGAUCGGAGGGCGCAAGUCCUGGACAGGAUCCGGCUCUGAAUCCGGGACUGAGUCCGGGUCCGGGUCCAAAUAUCC ............(((((((..(((((....)))))((((.(((..(((..(((....)))..)))..)))..)))).)))))))...... ( -38.80) >DroSec_CAF1 107291 90 - 1 GAUGAGAGUGAAUUGGAUCGGAGGGCGCAAGUCCUGGACAGGAUCCGGCUCUGAAUCCGGGACUGAGUCCGGGUCCGUGUCCAAAUAUCC (((..((.....))..)))(((((((....))))(((((((((((((((((.............))).)))))))).))))))....))) ( -38.92) >DroSim_CAF1 101050 78 - 1 GAUGAGAGUGAAUUGGAUCGGAGGGCGCAAGUCCUGGCCAGGAUCCGGCUCUG------------AGUGCGGGUCCGGGUCCAAAUAUCC ............((((((((((((((....)))))..)).(((((((((....------------.)).))))))).)))))))...... ( -32.80) >DroEre_CAF1 110969 84 - 1 GAUGAGAGUGAAUUGGAUCGGAGGGCGUAAGUCCUGGCCAGGAUCCGGCUCCGAGUCCGGCUCCU------AGUCCGGGUCCAAACAUCC ((((.........(((.((..(((((....))))))))))((((((((((..(((.....)))..------)).))))))))...)))). ( -34.60) >consensus GAUGAGAGUGAAUUGGAUCGGAGGGCGCAAGUCCUGGACAGGAUCCGGCUCUGAAUCCGGGACUGAGUCCGGGUCCGGGUCCAAAUAUCC ............(((((((..(((((....))))).....(((((((((((.............))).)))))))).)))))))...... (-26.17 = -28.67 + 2.50)


| Location | 17,223,890 – 17,224,010 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.67 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -33.78 |
| Energy contribution | -34.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17223890 120 + 23771897 UGUCCAGGACUUGCGCCCUCCGAUCCAAUUCACUCUCAUCCGCUAGUGGAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUUGGAUACCGUUGCUCCUGGCCAUUAAAAGUAUGC ((.((((((...((((((((((((..((((.((((.((((((....))))......))...)))))))).))).))))))((((...))))..)))...)))))).))............ ( -33.50) >DroSec_CAF1 107341 120 + 1 UGUCCAGGACUUGCGCCCUCCGAUCCAAUUCACUCUCAUCCGCUAGUGGAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUUGGAUACCGUUGCUCCUGGCCAUUAAAAGUAUGC ((.((((((...((((((((((((..((((.((((.((((((....))))......))...)))))))).))).))))))((((...))))..)))...)))))).))............ ( -33.50) >DroSim_CAF1 101088 120 + 1 UGGCCAGGACUUGCGCCCUCCGAUCCAAUUCACUCUCAUCCGCUAGUGGAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUUGGAUACCGUUGCUCCUGGCCAUUAAAAGUAUGC (((((((((...((((((((((((..((((.((((.((((((....))))......))...)))))))).))).))))))((((...))))..)))...)))))))))............ ( -40.60) >DroEre_CAF1 111013 120 + 1 UGGCCAGGACUUACGCCCUCCGAUCCAAUUCACUCUCAUCCGCUGGUGCAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUGGGAUACCGUAGCUCCUGGCCAUUAAAAGUAUGC (((((((((.(((((((((((((......))..........(((.((((((....)))))).))).........))))))((((...))))..))))).)))))))))............ ( -47.70) >DroYak_CAF1 113736 120 + 1 UGGCCAGGACUUACGCCCUGCGAUCCAAUUCACUAGCAUCCGCUAGUGCAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUUGGAUGCCGUAGCUCCUGGCCAUUAAAAGUACGC (((((((((.(((((((((((((.......(((((((....))))))).......))))).))))............((.((((...)))).)))))).)))))))))............ ( -51.54) >consensus UGGCCAGGACUUGCGCCCUCCGAUCCAAUUCACUCUCAUCCGCUAGUGGAAAAUUUUGCAUGGGUAAUUAAUCUGGAGGGAUCCCUUGGAUACCGUUGCUCCUGGCCAUUAAAAGUAUGC (((((((((...((((((((((((....((((((..(....)..)))))).....((((....))))...))).))))))((((...))))..)))...)))))))))............ (-33.78 = -34.38 + 0.60)

| Location | 17,224,010 – 17,224,130 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -29.52 |
| Consensus MFE | -29.00 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224010 120 + 23771897 AGCUCGGCAAUUUCUGAAAUUUUAAUAUGGCAAACGAACUAACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAA (((((((......))))........((((((....(((.((((..((((.......)))))))).))).....)))))))))...........(((((((((......)))))))))... ( -29.60) >DroSec_CAF1 107461 120 + 1 AGCUCGGCAAUUUCUGAAAUUUUAAUAUGGCAAACGAACUAACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCCAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAA (((..(((((((.....))))....((((((....(((.((((..((((.......)))))))).))).....))))))))).....)))...(((((((((......)))))))))... ( -29.30) >DroEre_CAF1 111133 120 + 1 AGCUCGGCAAUUUCUGAAAUUUUAAUAUGGCAAACGAACUAACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAA (((((((......))))........((((((....(((.((((..((((.......)))))))).))).....)))))))))...........(((((((((......)))))))))... ( -29.60) >DroYak_CAF1 113856 120 + 1 AGCUCGGCAAUUUCUGAAAUUUUAAUAUGGCAAACGAACUAACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAA (((((((......))))........((((((....(((.((((..((((.......)))))))).))).....)))))))))...........(((((((((......)))))))))... ( -29.60) >consensus AGCUCGGCAAUUUCUGAAAUUUUAAUAUGGCAAACGAACUAACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAA (((((((......))))........((((((....(((.((((..((((.......)))))))).))).....))))))........)))...(((((((((......)))))))))... (-29.00 = -29.00 + 0.00)


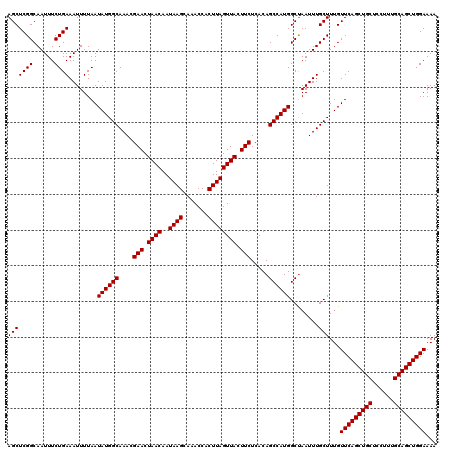
| Location | 17,224,050 – 17,224,166 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -33.01 |
| Energy contribution | -33.33 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224050 116 + 23771897 AACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUAGAUGC ........(((....((((((((((.......(((....))).((((.(....(((((((((......)))))))))....).))))((((.....)))))))))----)))))...))) ( -36.70) >DroSec_CAF1 107501 116 + 1 AACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCCAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUGGAUGC ........(((..((((((((((((.........(((((((......)))...(((((((((......)))))))))..))))....((((.....)))))))))----))))))).))) ( -41.80) >DroEre_CAF1 111173 116 + 1 AACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUGGAUGC ........(((..((((((((((((.......(((....))).((((.(....(((((((((......)))))))))....).))))((((.....)))))))))----))))))).))) ( -43.00) >DroYak_CAF1 113896 120 + 1 AACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUUUUCGUAGCCGGAUGGGUAGAUGC ........(((.(((....(((.(((((......(((((((......)))...(((((((((......)))))))))..))))....))))).))).((((....))))..)))...))) ( -33.10) >consensus AACAAUAAGCAAACCACUUAGUUACUUCUCACAGCCAUGGCUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC____UGGGUAGAUGC ........(((..((((((((((((.........(((((((......)))...(((((((((......)))))))))..))))....((((.....)))))))))....))))))).))) (-33.01 = -33.33 + 0.31)

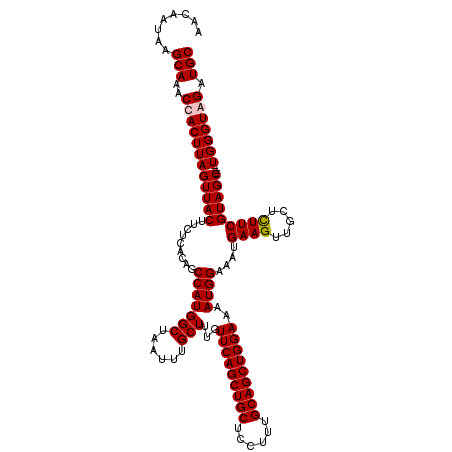

| Location | 17,224,050 – 17,224,166 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.89 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -32.24 |
| Energy contribution | -31.80 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224050 116 - 23771897 GCAUCUACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAGCCAUGGCUGUGAGAAGUAACUAAGUGGUUUGCUUAUUGUU (((.((((..(----(.(((((((.....)))).......((((((((((((......)))))))............((((....)))).)))))))).))..))))..)))........ ( -31.60) >DroSec_CAF1 107501 116 - 1 GCAUCCACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUGGCCAUGGCUGUGAGAAGUAACUAAGUGGUUUGCUUAUUGUU (((.((((..(----(.(((((((.....)))).......((((((((((((......))))))).............(((....)))..)))))))).))..))))..)))........ ( -33.00) >DroEre_CAF1 111173 116 - 1 GCAUCCACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAGCCAUGGCUGUGAGAAGUAACUAAGUGGUUUGCUUAUUGUU (((.((((..(----(.(((((((.....)))).......((((((((((((......)))))))............((((....)))).)))))))).))..))))..)))........ ( -33.90) >DroYak_CAF1 113896 120 - 1 GCAUCUACCCAUCCGGCUACGAAAAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAGCCAUGGCUGUGAGAAGUAACUAAGUGGUUUGCUUAUUGUU (((.((((.......(((......))).(((((............(((((((......)))))))............((((....))))...)))))......))))..)))........ ( -30.90) >consensus GCAUCCACCCA____GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAGCCAUGGCUGUGAGAAGUAACUAAGUGGUUUGCUUAUUGUU (((.((((.......(((......))).(((((............(((((((......)))))))............((((....))))...)))))......))))..)))........ (-32.24 = -31.80 + -0.44)



| Location | 17,224,090 – 17,224,206 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -33.12 |
| Consensus MFE | -28.37 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224090 116 + 23771897 CUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUAGAUGCAAGUUUUGGUAGCCACUUGAAAUAUGCAAAAUUUGUAAUA ...((((((((..(((((((((......)))))))))....((..((((((.....))))))..)----)))))))))(((((((((((((...........))).)))))))))).... ( -33.40) >DroSec_CAF1 107541 105 + 1 CCAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUGGAUGCCAGUUU-----------UGAAAUAUGCAAAAUUUGUAAUA (((....(((...(((((((((......)))))))))........((((((.....)))))))))----....))).(((.(((((-----------((.......))))))).)))... ( -30.90) >DroEre_CAF1 111213 116 + 1 CUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC----UGGGUGGAUGCAAGUUUUGGUAGCCACUUGAAAUAUGCAAAAUUUGUAAUA ....(((((....(((((((((......)))))))))....((..((((((.....))))))..)----)((((((.(((........))).)))))).......))))).......... ( -33.90) >DroYak_CAF1 113936 120 + 1 CUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUUUUCGUAGCCGGAUGGGUAGAUGCAAGUUUUGGUAGCCACUUGAAAUAUGCAAAAUUUGUAAUA ...((((((((..(((((((((......)))))))))...(((..((((((.....))))))..)))...))))))))(((((((((((((...........))).)))))))))).... ( -34.30) >consensus CUAAUUUGCUUUGUUCAGCUGCUCCUUUGCAGCUGGAAAAUGGAAAUGAAGUUGCUCUUCGUAGC____UGGGUAGAUGCAAGUUUUGGUAGCCACUUGAAAUAUGCAAAAUUUGUAAUA (((....(((...(((((((((......)))))))))........((((((.....)))))))))....))).....(((((((((((..................)))))))))))... (-28.37 = -28.74 + 0.37)

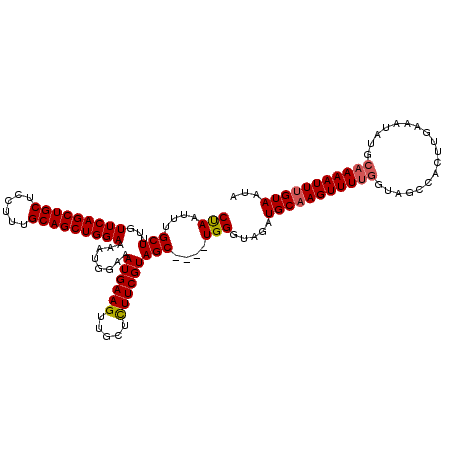
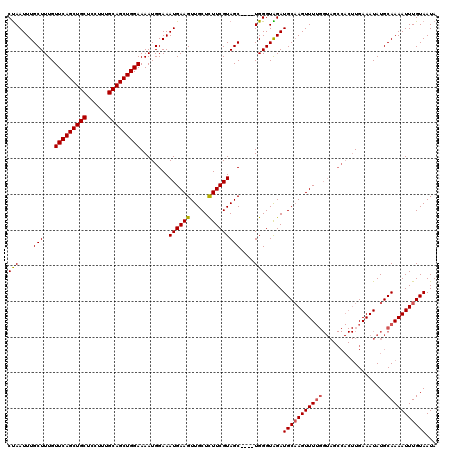
| Location | 17,224,090 – 17,224,206 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -24.94 |
| Consensus MFE | -19.98 |
| Energy contribution | -19.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17224090 116 - 23771897 UAUUACAAAUUUUGCAUAUUUCAAGUGGCUACCAAAACUUGCAUCUACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAG ..........(((((.........((((((....................)----)))))((((.....))))............(((((((......)))))))......))))).... ( -25.85) >DroSec_CAF1 107541 105 - 1 UAUUACAAAUUUUGCAUAUUUCA-----------AAACUGGCAUCCACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUGG .....(((..(((((........-----------...((((.......)))----)....((((.....))))............(((((((......)))))))......)))))))). ( -24.00) >DroEre_CAF1 111213 116 - 1 UAUUACAAAUUUUGCAUAUUUCAAGUGGCUACCAAAACUUGCAUCCACCCA----GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAG ..........(((((.........((((((....................)----)))))((((.....))))............(((((((......)))))))......))))).... ( -25.85) >DroYak_CAF1 113936 120 - 1 UAUUACAAAUUUUGCAUAUUUCAAGUGGCUACCAAAACUUGCAUCUACCCAUCCGGCUACGAAAAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAG ..........(((((.......((((((..........((((.((((.((....)).)).))...))))........))))))..(((((((......)))))))......))))).... ( -24.07) >consensus UAUUACAAAUUUUGCAUAUUUCAAGUGGCUACCAAAACUUGCAUCCACCCA____GCUACGAAGAGCAACUUCAUUUCCAUUUUCCAGCUGCAAAGGAGCAGCUGAACAAAGCAAAUUAG ..........(((((........................................(((......)))..................(((((((......)))))))......))))).... (-19.98 = -19.98 + -0.00)

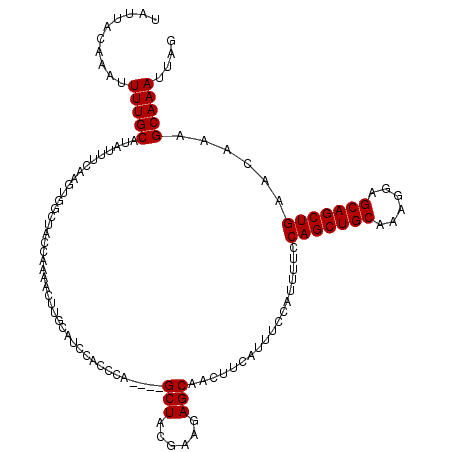
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:53 2006