
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,223,171 – 17,223,321 |
| Length | 150 |
| Max. P | 0.991565 |

| Location | 17,223,171 – 17,223,289 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -49.06 |
| Consensus MFE | -37.34 |
| Energy contribution | -38.14 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17223171 118 + 23771897 -CGGG-ACCCCAACAUUGCCCACAUCGCUGCGAGGAUUGGGGAAGUCUGCUUGUGGCAGGGGUUUUGGCCAGUCGGGGGCCGUGGGCUCAUUAUGUUGGGGCUUUUUAAUUAUGGCCAAA -.(((-.(((((((((.((((((...((..((((((((.....))))..))))..)).........((((.......)))))))))).....))))))))))))................ ( -48.00) >DroSec_CAF1 106706 119 + 1 -CGGGAACCCCAGCAUUGCCCACAUCGCUGCGGGGAUUGGGGAAGUCUGCUUGUGGCGGGGGUUUUGGCCAGUCGGGGGCCGUGGGCUCAUUAUGUUGGGGCUUUUUAAUUAUGGCCAAA -.((((((((((((((.((((((.((((..((((((((.....))))..))))..)))).......((((.......)))))))))).....))))))))).)))))............. ( -49.70) >DroSim_CAF1 100447 119 + 1 -CGGGAACCCCAGCAUUGCCCACAUCGCUGCGGGGAUUGGGGAAGUCUGCUUGUGGCAGGGGUUUUGGCCAGUCGGGGGCCGUGGGCUCAUUAUGUUGGGGCUUUUUAAUUAUGGCCAAA -.((((((((((((((.(((((((((.(((((.(((((.....))))).).....)))).)))...((((.......)))))))))).....))))))))).)))))............. ( -47.40) >DroEre_CAF1 110251 116 + 1 -UGGGA-CCCCAACAUUGCCCACAUCCCUGCG--GAUUGGGGUAUUCUGCUUUUGGCAGGGGUUUUGGCCAGUCGGGGGCGGUGGGCUCAUUAUGUUGGGGCAUUUUAAUUAUGGCCAAA -(((..-(((((((((.((((((((((((((.--((..(.((....)).)..)).))))))))....(((.......))).)))))).....)))))))))(((.......))).))).. ( -51.60) >DroYak_CAF1 112917 118 + 1 UUGGGAGCCCCAACUUUGCCCACAUCCCUGCG--GAUUGGGGUAGUCUGCAUUUGGCAGGGGUUUUGGCCAGUCGCUCGCUGUGAGCUCAUUAUGUUGGGGCUUUUUAAUUAUGGCCAAA ((((((((((((((...(((...((((((((.--((.((.((....)).)).)).))))))))...))).....((((.....)))).......))))))))))...........)))). ( -48.60) >consensus _CGGGAACCCCAACAUUGCCCACAUCGCUGCG_GGAUUGGGGAAGUCUGCUUGUGGCAGGGGUUUUGGCCAGUCGGGGGCCGUGGGCUCAUUAUGUUGGGGCUUUUUAAUUAUGGCCAAA .......(((((((((.(((((((((.((((.(.((.((((....)))).)).).)))).)))...((((.......)))))))))).....)))))))))................... (-37.34 = -38.14 + 0.80)

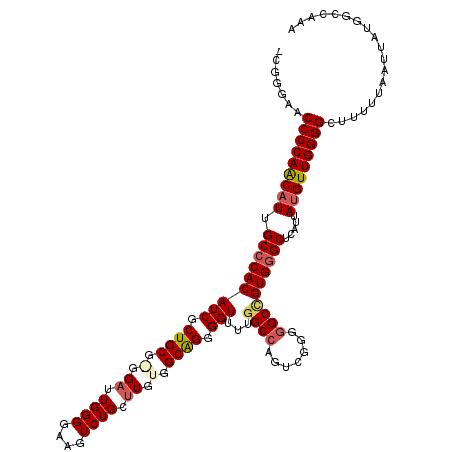
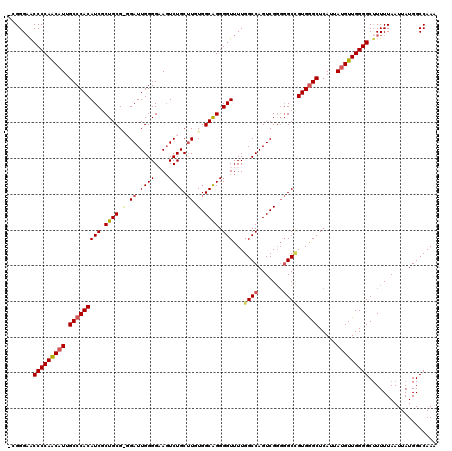
| Location | 17,223,171 – 17,223,289 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -30.58 |
| Energy contribution | -31.90 |
| Covariance contribution | 1.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17223171 118 - 23771897 UUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCCCCAAUCCUCGCAGCGAUGUGGGCAAUGUUGGGGU-CCCG- ..................((((((((((.....((((((((((.......)))).......((((.....)))).......................)))))).))))))))))-....- ( -38.70) >DroSec_CAF1 106706 119 - 1 UUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCCGCCACAAGCAGACUUCCCCAAUCCCCGCAGCGAUGUGGGCAAUGCUGGGGUUCCCG- .(((((((..........((..((......)).))...(((...)))..)))))))((((((.((...(((....)))........((((((....))))))....)).))))))....- ( -31.80) >DroSim_CAF1 100447 119 - 1 UUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCCCCAAUCCCCGCAGCGAUGUGGGCAAUGCUGGGGUUCCCG- .................(((((((.(((.....((((((((((.......)))).......((((.....)))).......................)))))).))).)))))))....- ( -35.10) >DroEre_CAF1 110251 116 - 1 UUUGGCCAUAAUUAAAAUGCCCCAACAUAAUGAGCCCACCGCCCCCGACUGGCCAAAACCCCUGCCAAAAGCAGAAUACCCCAAUC--CGCAGGGAUGUGGGCAAUGUUGGGG-UCCCA- ..................((((((((((.....((((((.(((.......)))......((((((.....................--.))))))..)))))).)))))))))-)....- ( -38.25) >DroYak_CAF1 112917 118 - 1 UUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCUCACAGCGAGCGACUGGCCAAAACCCCUGCCAAAUGCAGACUACCCCAAUC--CGCAGGGAUGUGGGCAAAGUUGGGGCUCCCAA .................(((((((((.......((((.....))))...((.(((....((((((....((..(....)..))...--.))))))...))).))..)))))))))..... ( -39.00) >consensus UUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCCCCAAUCC_CGCAGCGAUGUGGGCAAUGUUGGGGUUCCCG_ ..................((((((((((.....((((((((((.......)))).......((((.....)))).......................)))))).))))))))))...... (-30.58 = -31.90 + 1.32)

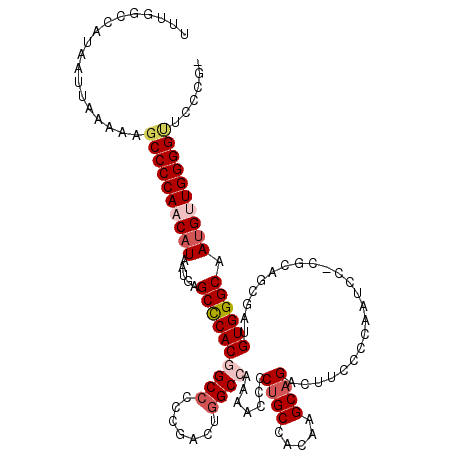
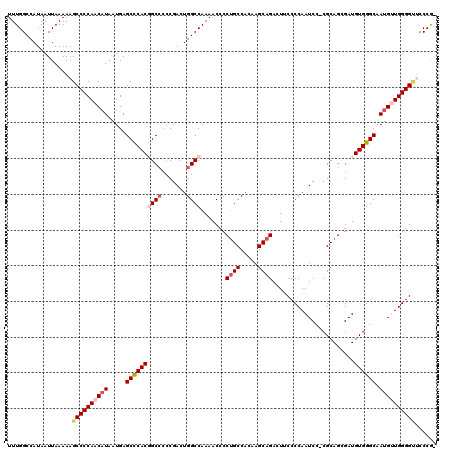
| Location | 17,223,209 – 17,223,321 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -21.56 |
| Consensus MFE | -18.86 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17223209 112 - 23771897 A--------AAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCC .--------.............((((........)))).(((((((((..........((..((......)).))...(((...)))..)))))))))...((((.....))))...... ( -21.60) >DroSec_CAF1 106745 112 - 1 A--------AAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCCGCCACAAGCAGACUUCC .--------..................((((....((..(((((((((..........((..((......)).))...(((...)))..))))))))).....)).....))))...... ( -19.10) >DroSim_CAF1 100486 112 - 1 A--------AAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCC .--------.............((((........)))).(((((((((..........((..((......)).))...(((...)))..)))))))))...((((.....))))...... ( -21.60) >DroEre_CAF1 110287 112 - 1 C--------AAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAUGCCCCAACAUAAUGAGCCCACCGCCCCCGACUGGCCAAAACCCCUGCCAAAAGCAGAAUACC .--------.............((((........)))).(((((((((........(((......))).....((.....)).......)))))))))...((((.....))))...... ( -19.40) >DroYak_CAF1 112955 120 - 1 AAAAGCAGAAAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCUCACAGCGAGCGACUGGCCAAAACCCCUGCCAAAUGCAGACUACC ....((..(((((...)))))..))..(((((...((..(((((((((..............((......)).((((.....))))...))))))))).....))....)))))...... ( -26.10) >consensus A________AAAUUUCAUUUUAAGCGACUGCAAACGCUCUUUUGGCCAUAAUUAAAAAGCCCCAACAUAAUGAGCCCACGGCCCCCGACUGGCCAAAACCCCUGCCACAAGCAGACUUCC ......................((((........)))).(((((((((..............((......)).((.....)).......)))))))))...((((.....))))...... (-18.86 = -19.06 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:47 2006