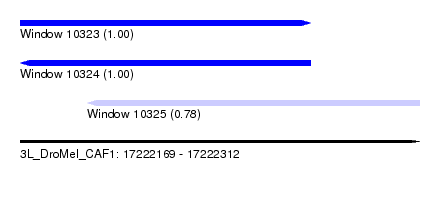
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 17,222,169 – 17,222,312 |
| Length | 143 |
| Max. P | 0.999934 |

| Location | 17,222,169 – 17,222,273 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -35.22 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
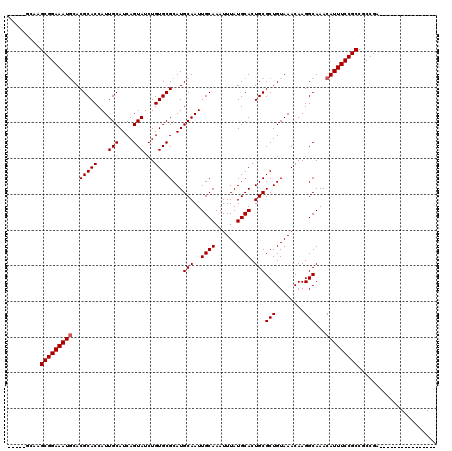
>3L_DroMel_CAF1 17222169 104 + 23771897 ----------------UCGGCGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGCAUUUCCGCUUGCUACUA ----------------..((((((((((((((.(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).))).)))))))))).)))).... ( -39.30) >DroSec_CAF1 105744 96 + 1 ----------------UCGGCGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGUAUUUCCGCC-------- ----------------.....((((((((((..(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).)))..))))))))))-------- ( -34.20) >DroSim_CAF1 99484 96 + 1 ----------------UCGGCGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGCAUUUCCGCU-------- ----------------.....(((((((((((.(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).))).)))))))))))-------- ( -35.50) >DroEre_CAF1 109310 100 + 1 ----------------UCGGCGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGCAUUUCCGCUUGCC---- ----------------..((((((((((((((.(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).))).)))))))))).))))---- ( -40.30) >DroYak_CAF1 111925 116 + 1 GCGAAUACUCGAAUACUCGGUGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGCAUUUCCGCUUGCU---- ((((.(((.(((....))))))((((((((((.(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).))).)))))))))))))).---- ( -40.90) >consensus ________________UCGGCGGCGGAAAUGUUUGCCUUGUUUACAGCGCAGUGCAUAAAUUUGCAAUUGCAUGCGCACAGAUACUGAUGCAAUGGUGCGUGCAUUUCCGCUUGC_____ .....................(((((((((((.(((.(..((....((((((((((......))).)))))..))((((((...))).)))))..).))).)))))))))))........ (-35.22 = -34.90 + -0.32)

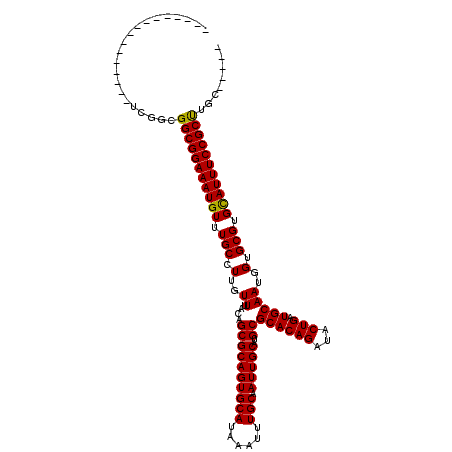

| Location | 17,222,169 – 17,222,273 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.03 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -29.56 |
| Energy contribution | -29.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17222169 104 - 23771897 UAGUAGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCGCCGA---------------- .....((..(((((((((..(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))...))))))))).))...---------------- ( -31.60) >DroSec_CAF1 105744 96 - 1 --------GGCGGAAAUACACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCGCCGA---------------- --------(((((((((...(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))....))))))))).....---------------- ( -32.00) >DroSim_CAF1 99484 96 - 1 --------AGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCGCCGA---------------- --------.(((((((((..(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))...)))))))))......---------------- ( -30.50) >DroEre_CAF1 109310 100 - 1 ----GGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCGCCGA---------------- ----(((..(((((((((..(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))...))))))))).)))..---------------- ( -35.60) >DroYak_CAF1 111925 116 - 1 ----AGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCACCGAGUAUUCGAGUAUUCGC ----.....(((((((((..(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))...)))))))))...(((((((....))))))). ( -35.70) >consensus _____GCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAACAAGGCAAACAUUUCCGCCGCCGA________________ .........(((((((((..(((((...(((....)))...)))))...(((..((((......)))).)))(((........)))...)))))))))...................... (-29.56 = -29.76 + 0.20)

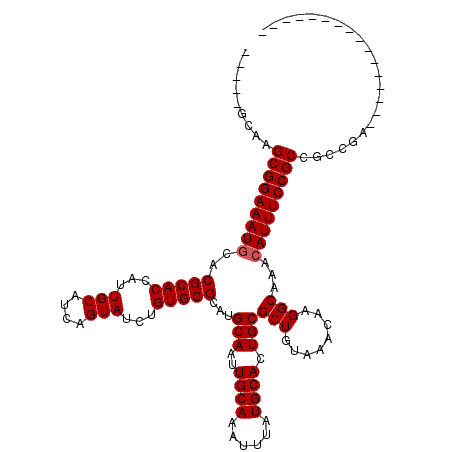
| Location | 17,222,193 – 17,222,312 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.32 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 17222193 119 - 23771897 GCGCCUCUU-CACACACCCACACACCUACACAGCCGCAUAUAGUAGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .........-...................(((((.((........))..((((...(((((((((...(((....)))...)))))..))))..((((......)))))))))))))... ( -29.00) >DroSec_CAF1 105768 104 - 1 GCCCCUCUU-CACACAC------ACGCACACACCCACAC---------GGCGGAAAUACACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .........-.......------..............((---------(((.........(((((...(((....)))...)))))...(((..((((......)))).))))))))... ( -21.40) >DroSim_CAF1 99508 104 - 1 GCCCCUCUU-CACACAC------ACGAACACAGCCACAC---------AGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .........-.......------......(((((.....---------.((((...(((((((((...(((....)))...)))))..))))..((((......)))))))))))))... ( -24.90) >DroEre_CAF1 109334 109 - 1 GCCCCCUUUUUCCACAC------ACACCCACACCCACAC-----GGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .................------..............((-----(((..((((...(((((((((...(((....)))...)))))..))))..((((......)))))))))))))... ( -25.80) >DroYak_CAF1 111965 102 - 1 GCCCCCUUU-------C------ACACACGCACCCACAC-----AGCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .........-------.------..............((-----(((..((((...(((((((((...(((....)))...)))))..))))..((((......)))))))))))))... ( -26.50) >consensus GCCCCUCUU_CACACAC______ACACACACACCCACAC______GCAAGCGGAAAUGCACGCACCAUUGCAUCAGUAUCUGUGCGCAUGCAAUUGCAAAUUUAUGCACUGCGCUGUAAA .................................................((((...(((((((((...(((....)))...)))))..))))..((((......))))))))........ (-19.56 = -19.76 + 0.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:18:41 2006